About us

Bin Liu's lab at Beijing Institute of Technology (BIT) is focusing on developing techniques grounded in the natural language processing (NLP) to uncover the meanings of "book of life". The research areas of Bin Liu's lab include:
1) Developing the Biological language models (BLMs);
2) Studying the natural language processing techniques;
3) Applying BLMs to biological sequence analysis;
4) Protein remote homology detection and fold recognition;
5) Predicting DNA/RNA binding proteins and their binding residues;
6) Disordered protein/region prediction based on sequence labelling models;
7) Predicting noncoding RNA-disease associations;
8) Identifying protein complexes;
9) DNA/RNA sequence analysis.
Web servers
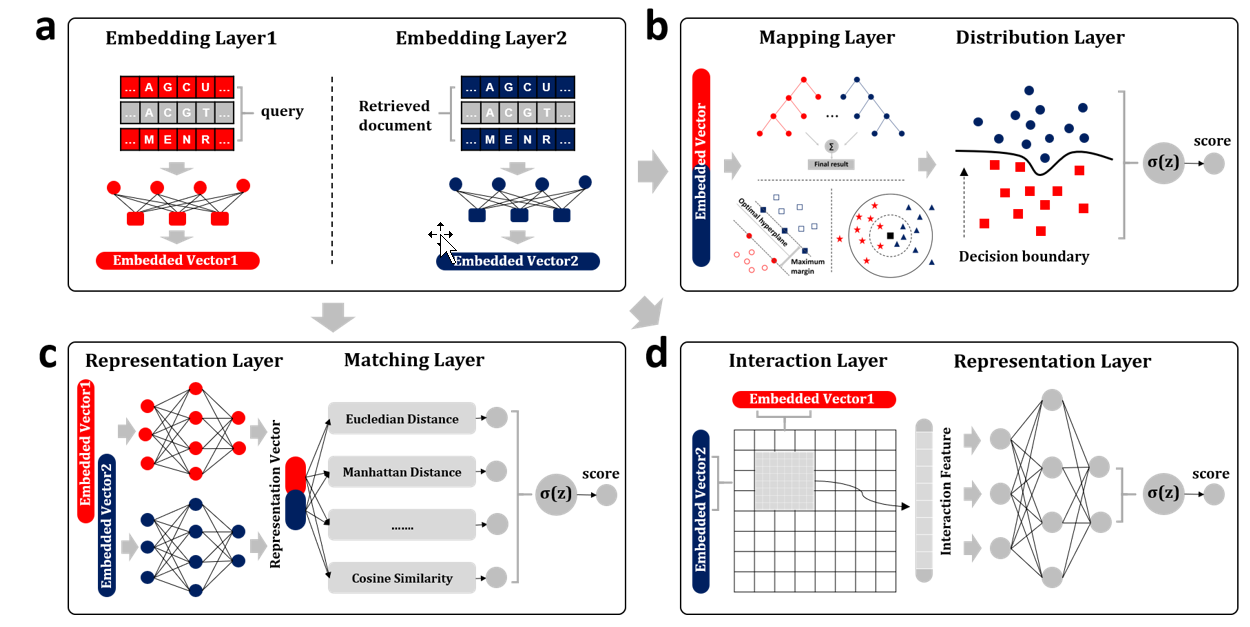
BioSeq-BLM
a platform for analyzing DNA, RNA, and protein sequences based on biological language models
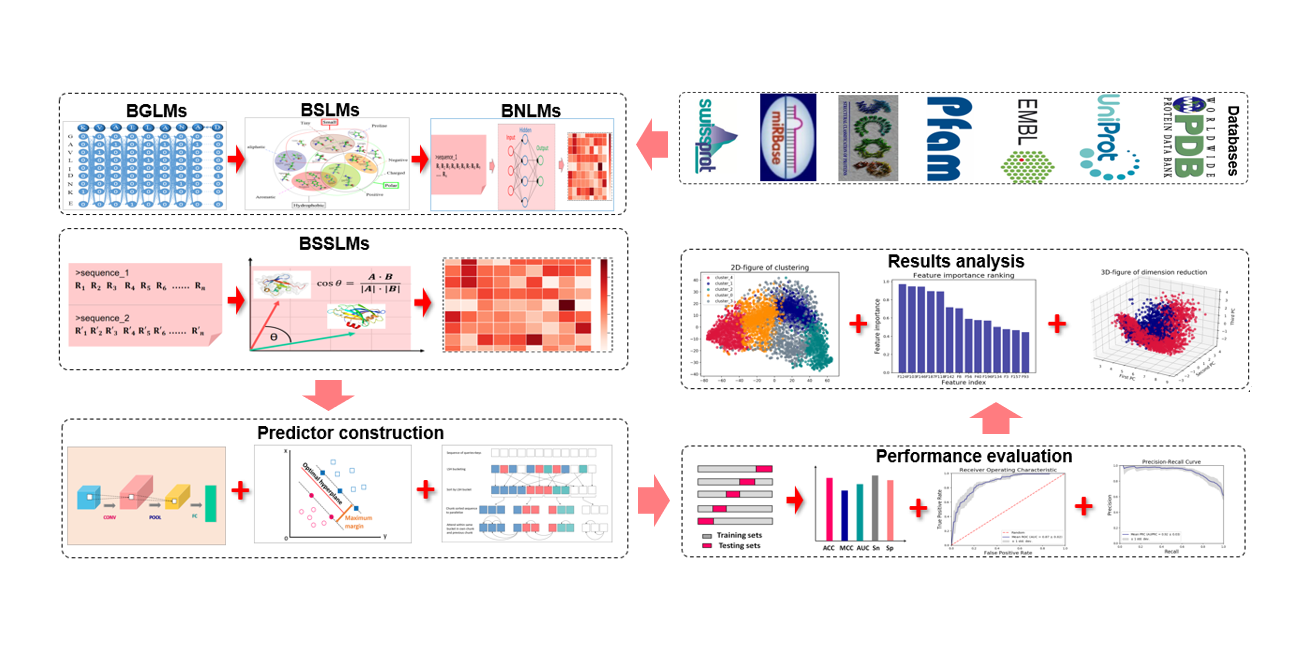
BioSeq-Analysis2.0
An updated platform for analyzing DNA, RNA and protein sequences at sequence level and residue level based ...
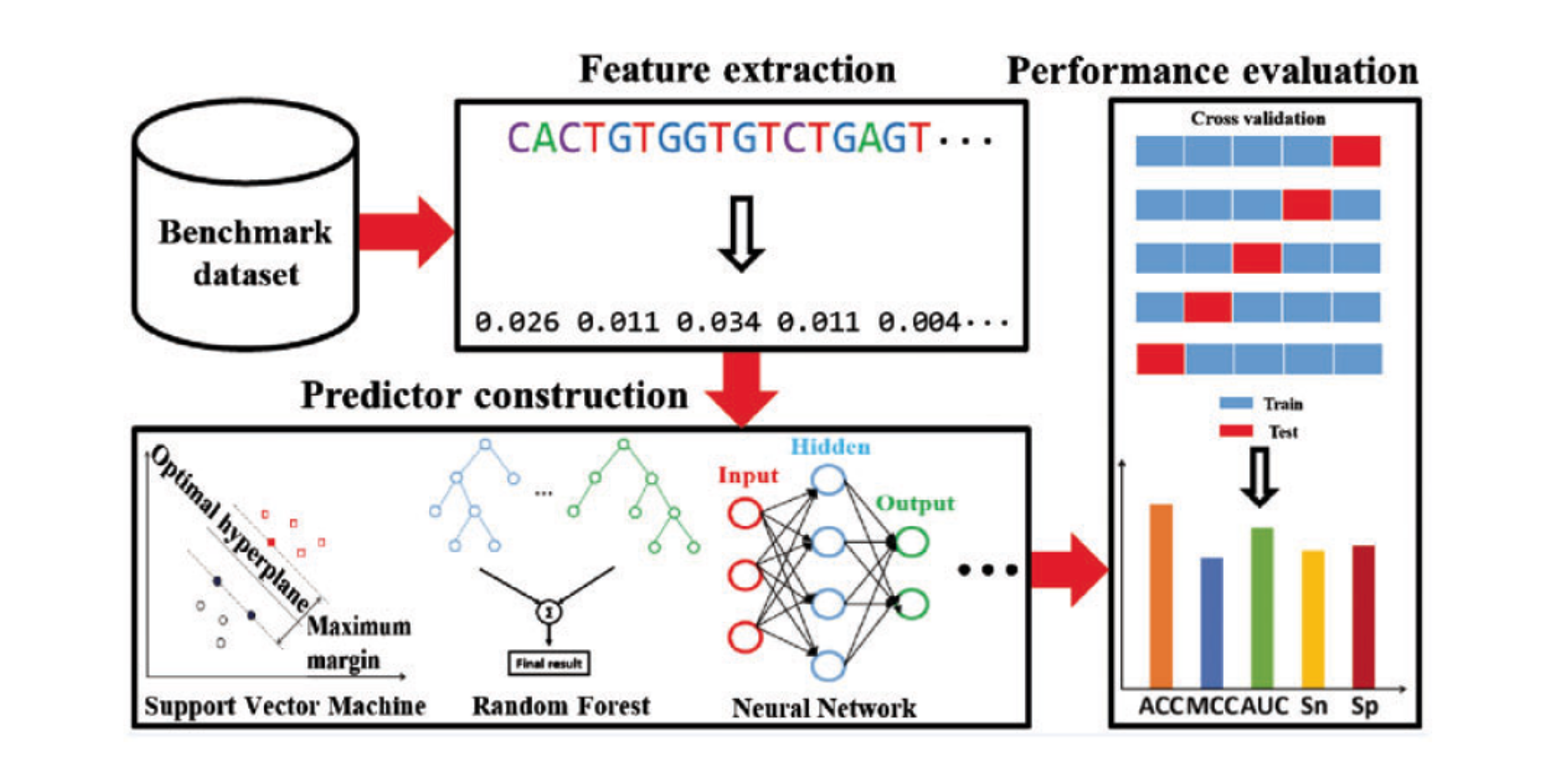
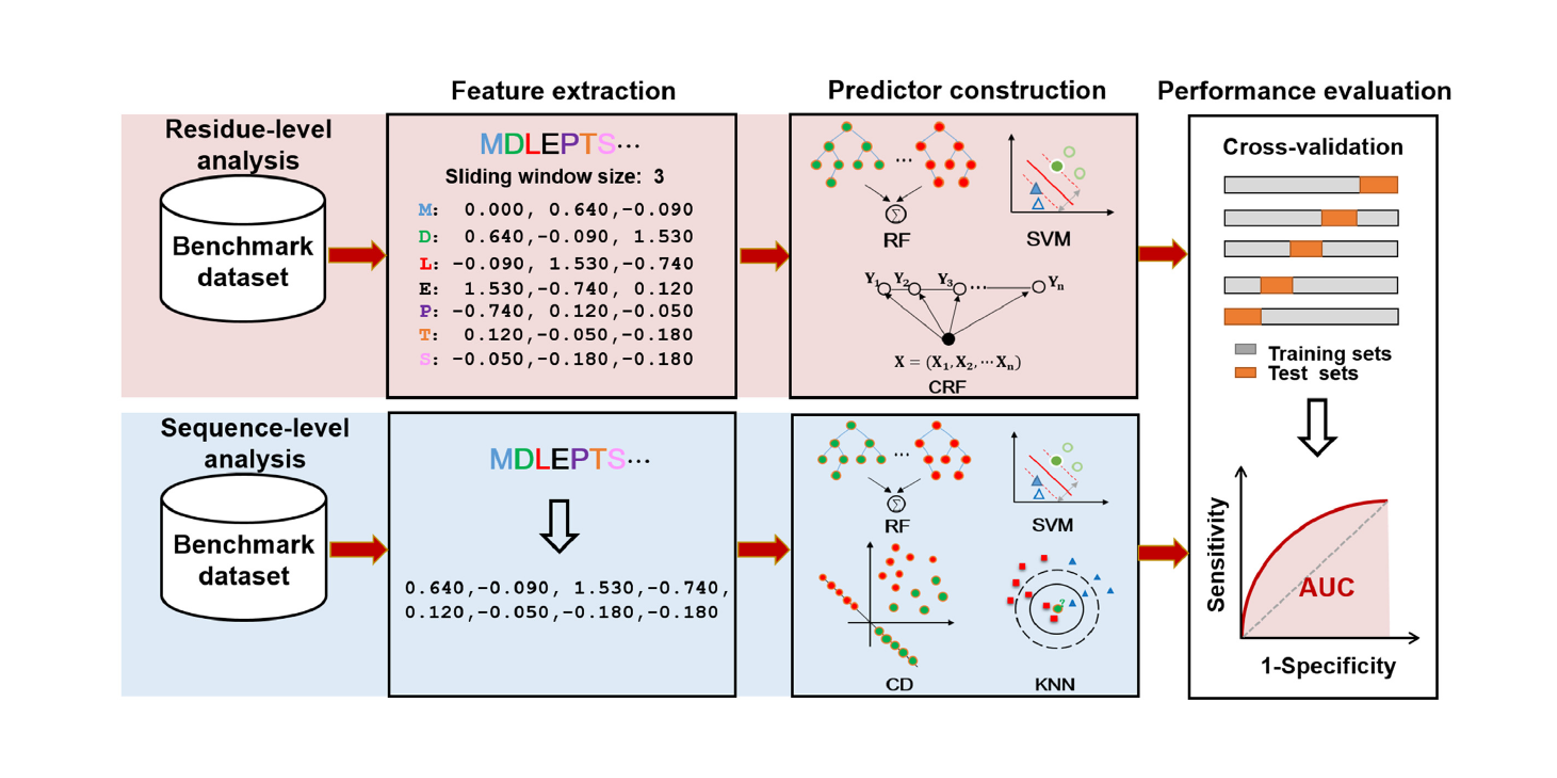
BioSeq-Analysis
A platform for DNA, RNA and protein sequence analysis based on machine learning approaches
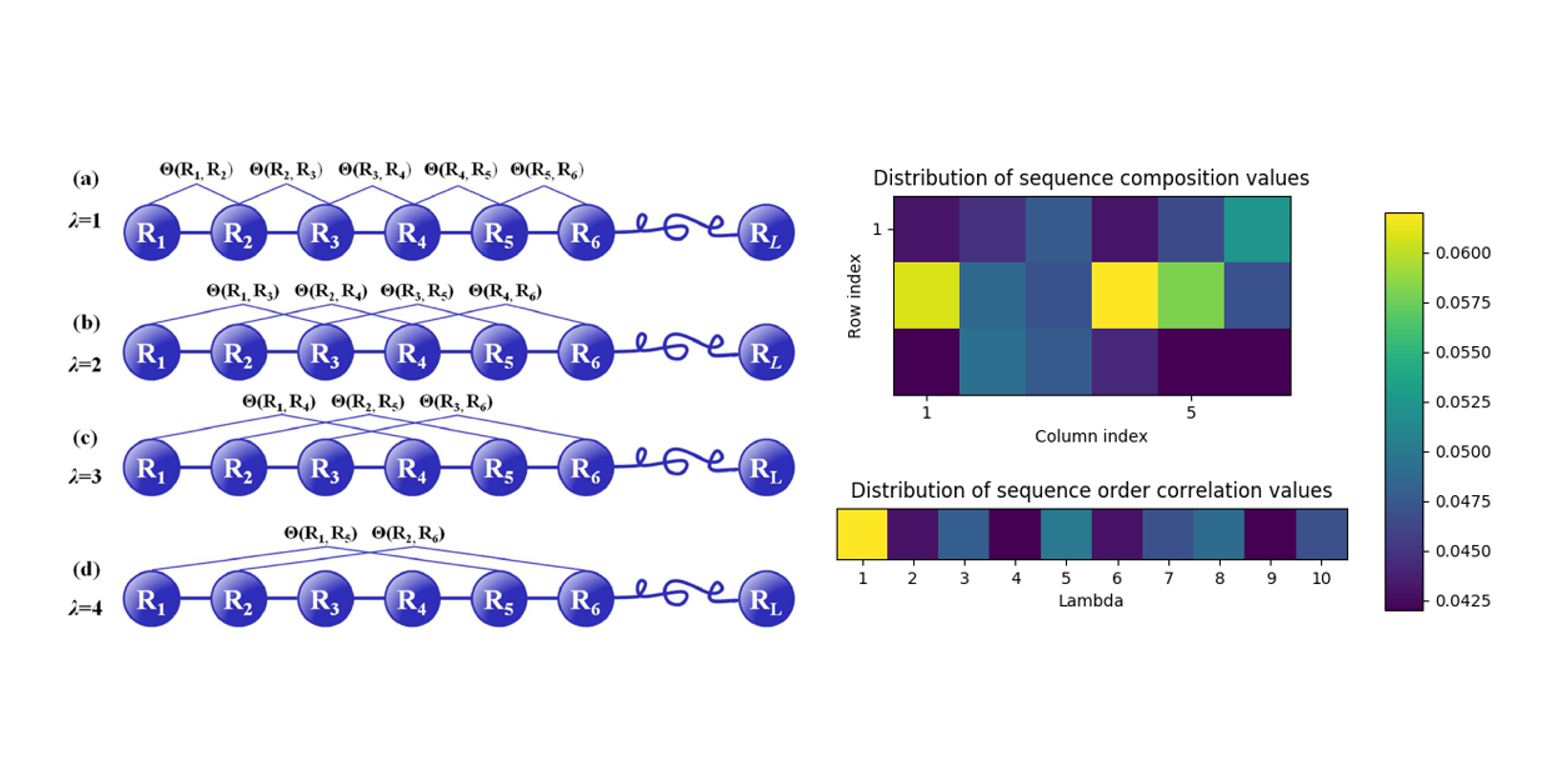
Pse-in-One
A web server for generating various modes of pseduo components of DNA, RNA, and protein sequences
Pse-Analysis
A Python package for DNA/RNA and protein/peptide sequence analysis based on pseudo components and kernel methods
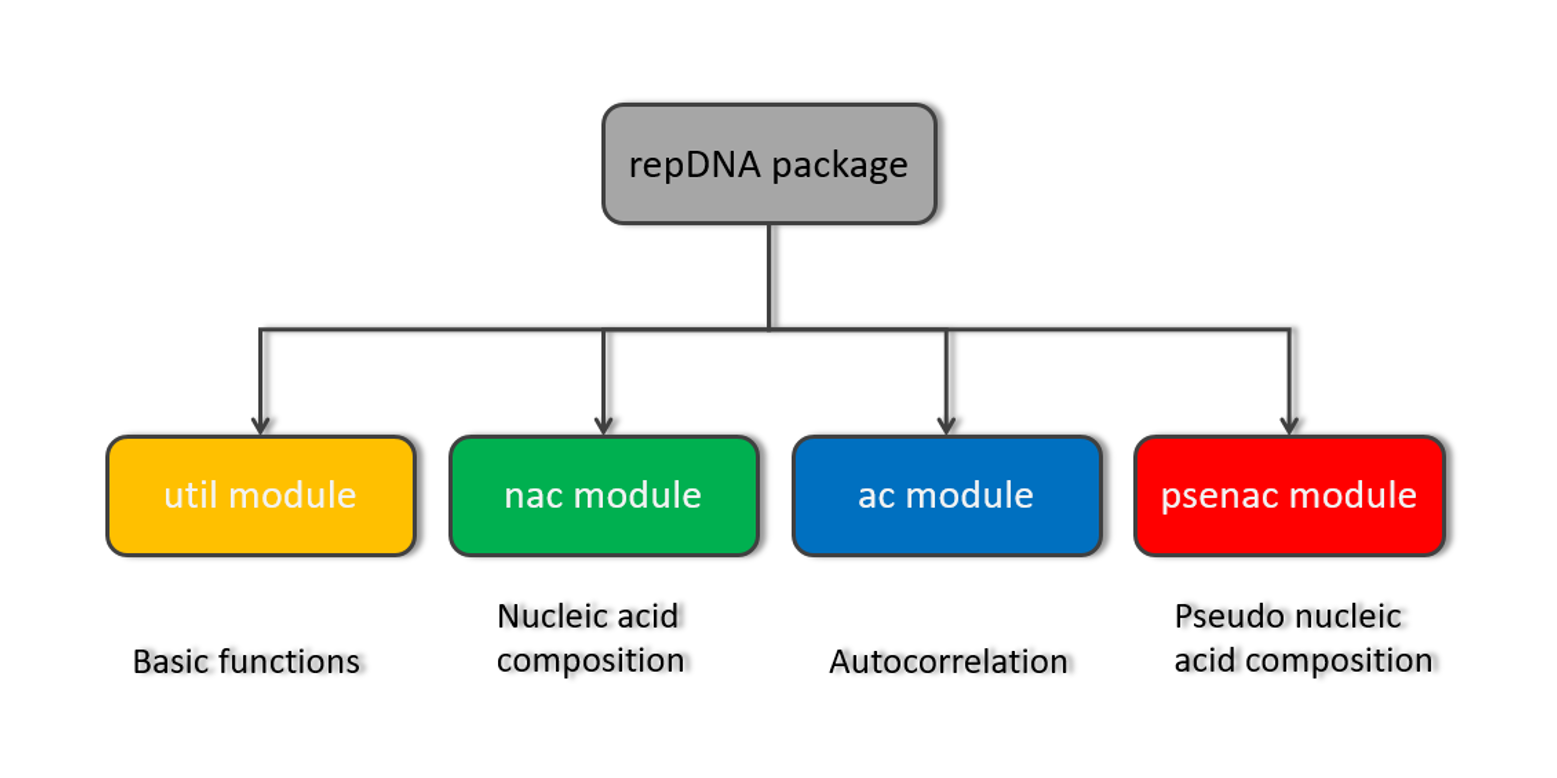
repDNA
A Python package to generate various modes of feature vectors for DNA sequences by incorporating user-defined ...
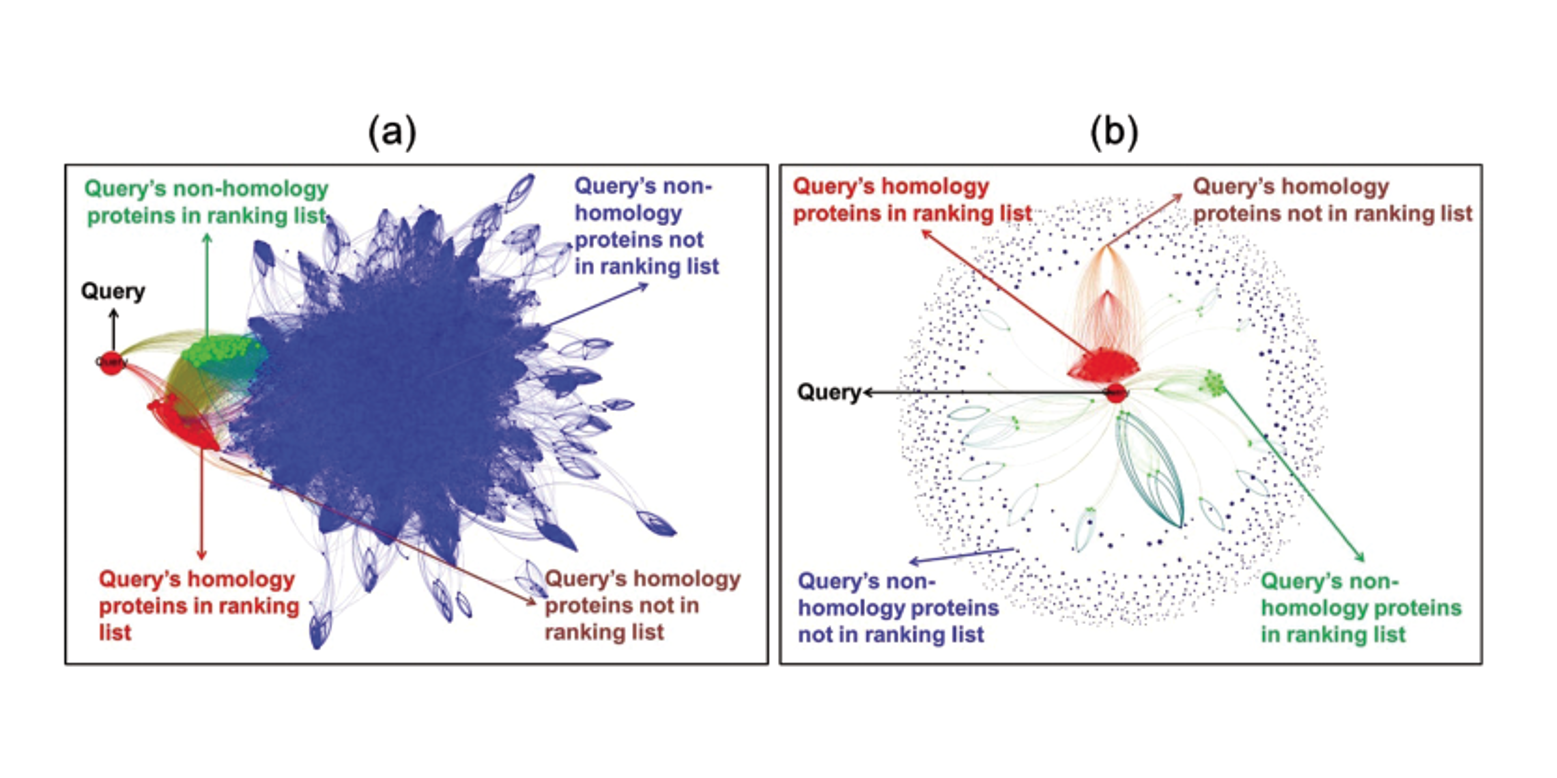
HITS-PR-HHblits
Protein Remote Homology Detection by Combining PageRank and Hyperlink-Induced Topic Search
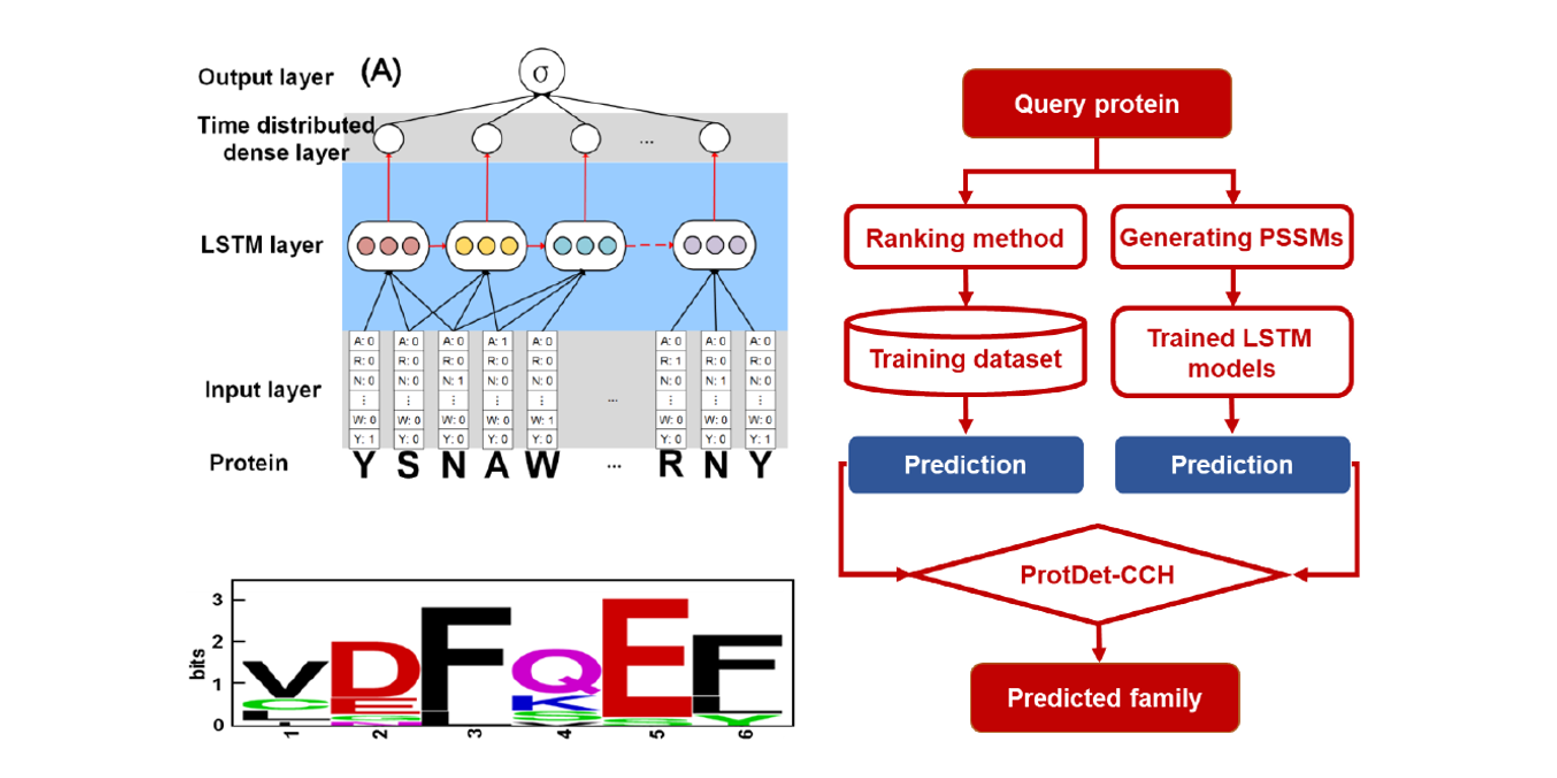
ProtDet-CCH
Protein remote homology detection by combining Long Short-Term Memory and ranking methods
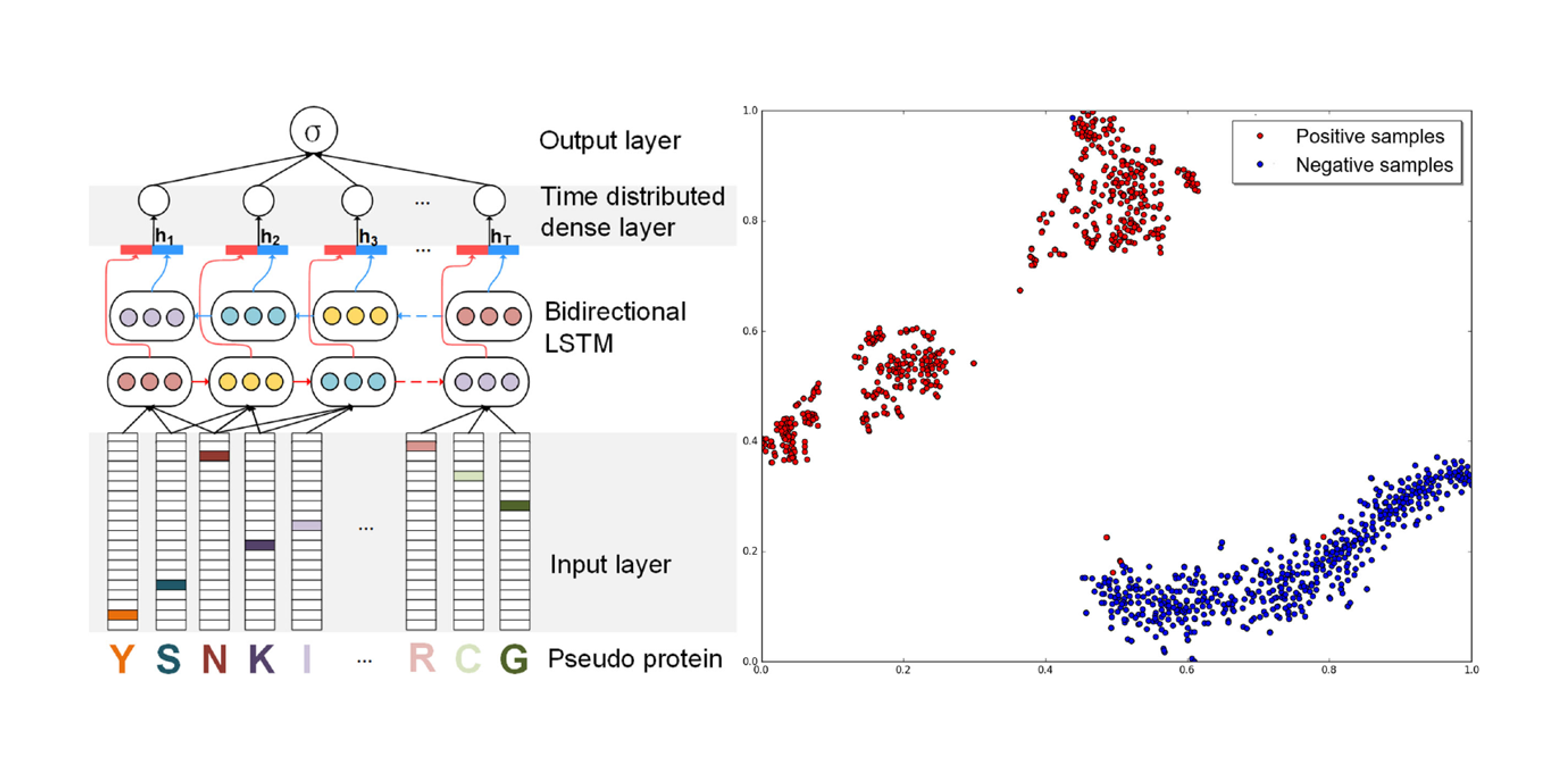
ProDec-BLSTM
Protein Remote Homology Detection based on Bidirectional Long Short-Term Memory
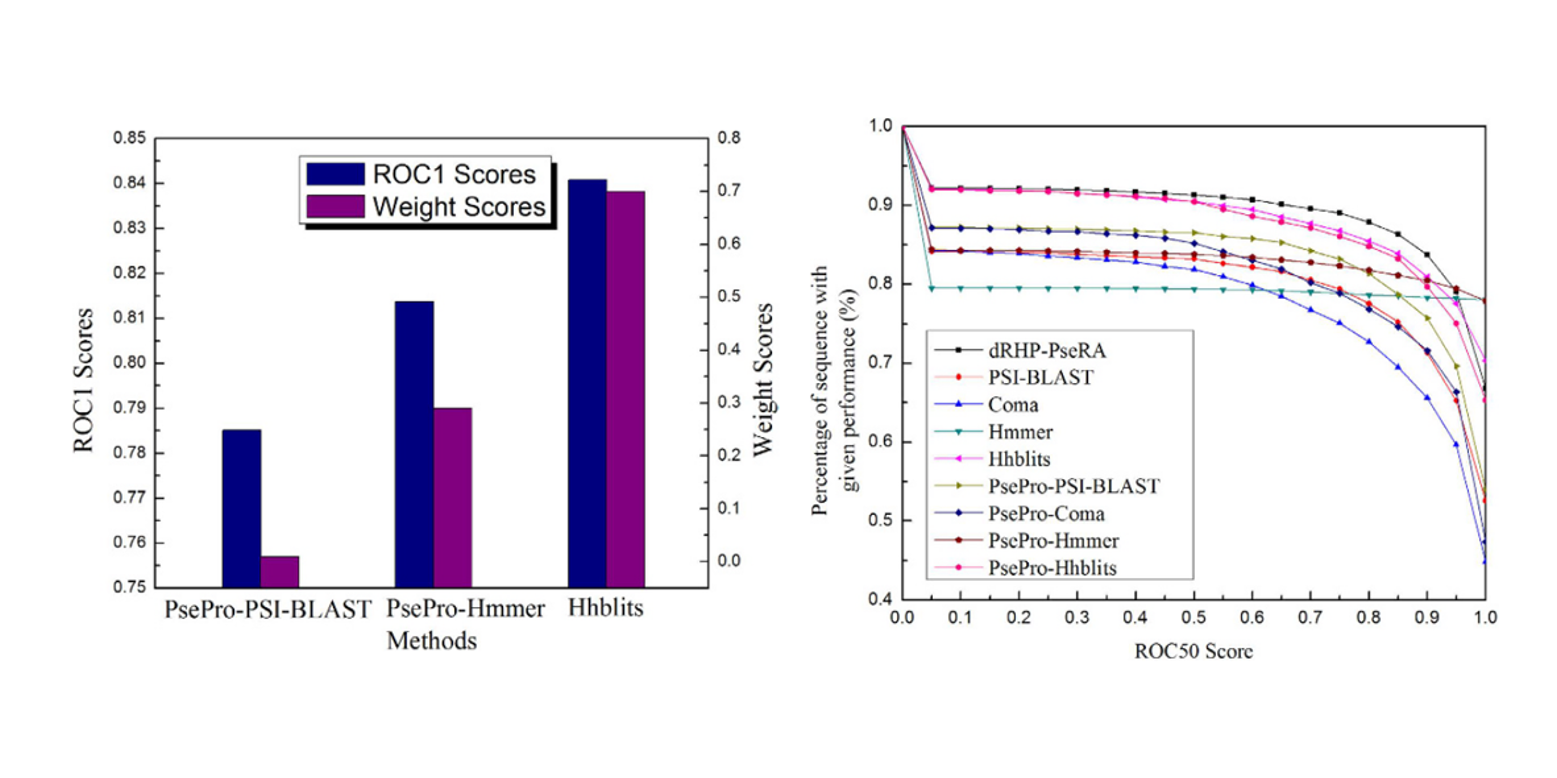
dRHP-PseRA
detecting remote homology proteins using profile-based pseudo protein sequence and rank aggregation
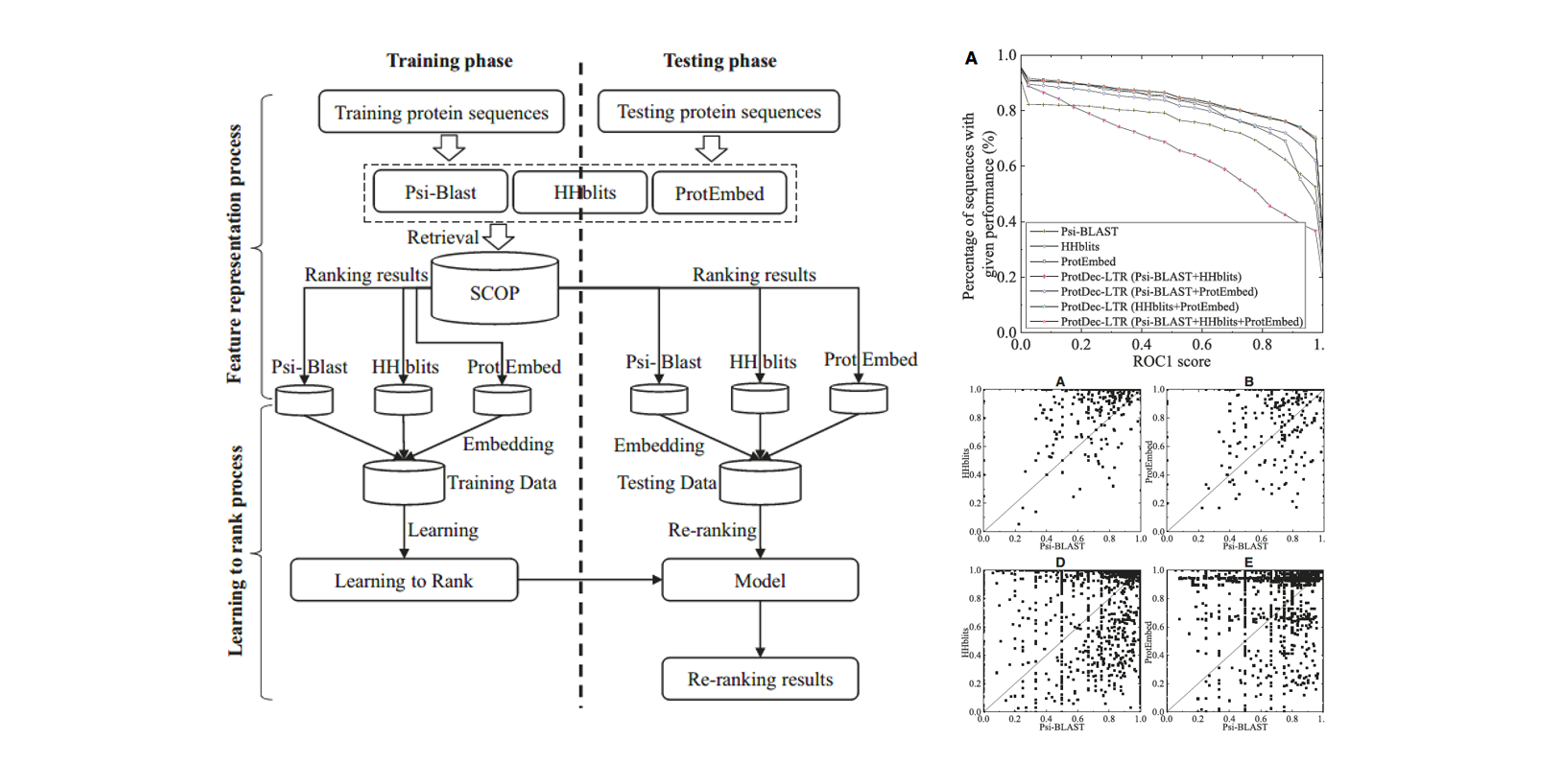
ProtDec-LTR
Application of Learning to Rank to protein remote homology detection
ProtDec-LTR2.0
An improved method for protein remote homology detection by combining pseudo protein and supervised learning to rank

ProtDec-LTR3.0
protein remote homology detection by incorporating profile-based features into Learning to Rank
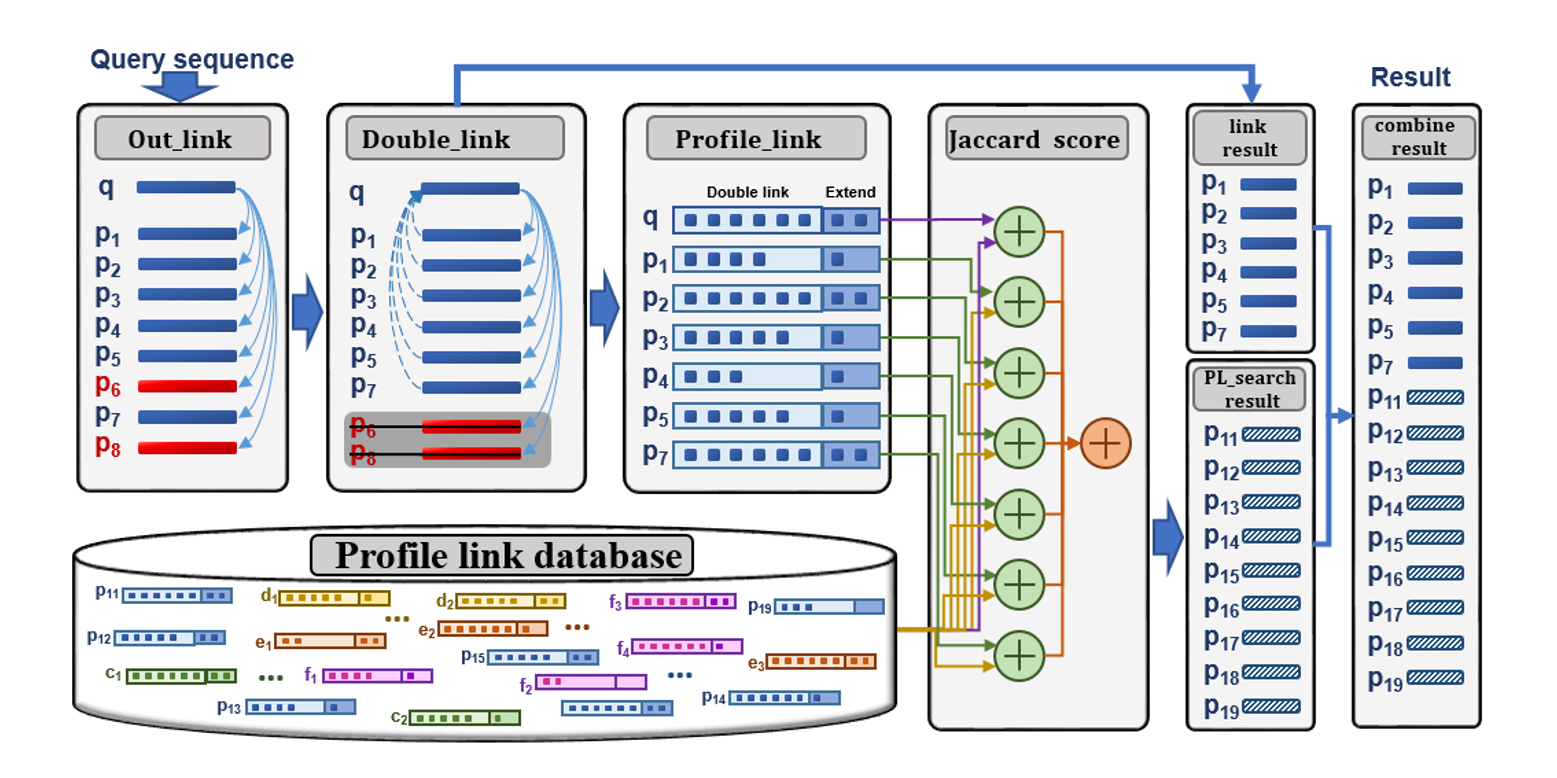
PL-search
A profile link based search method for protein remote homology detection
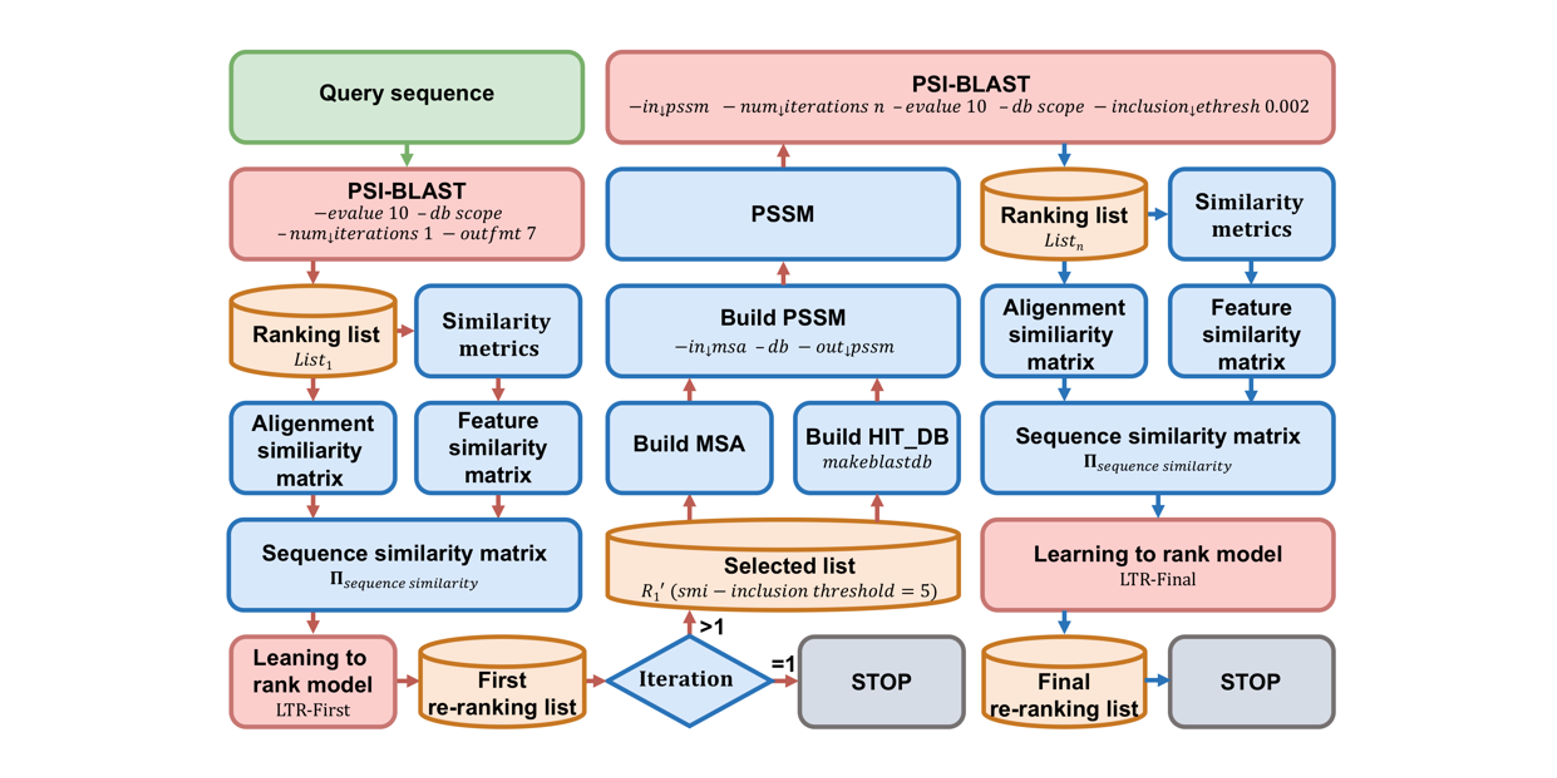
SMI-BLAST
A novel supervised search framework based on PSI-BLAST for protein remote homology detection and its application to ...
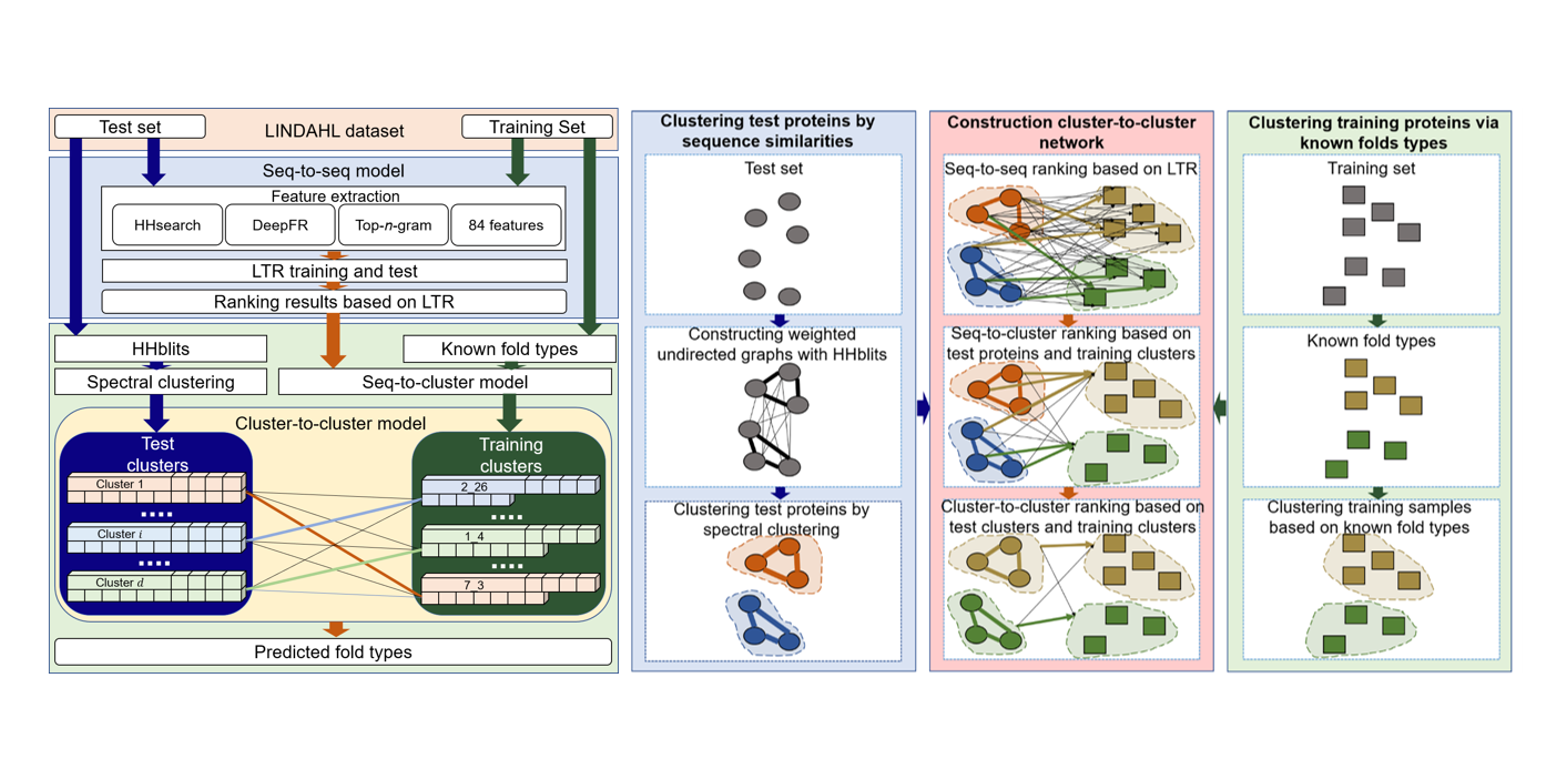
FoldRec-C2C
protein fold recognition by combining cluster-to-cluster model and protein similarity network

ProtFold-DFG
protein fold recognition by combining Directed Fusion Graph and PageRank algorithm
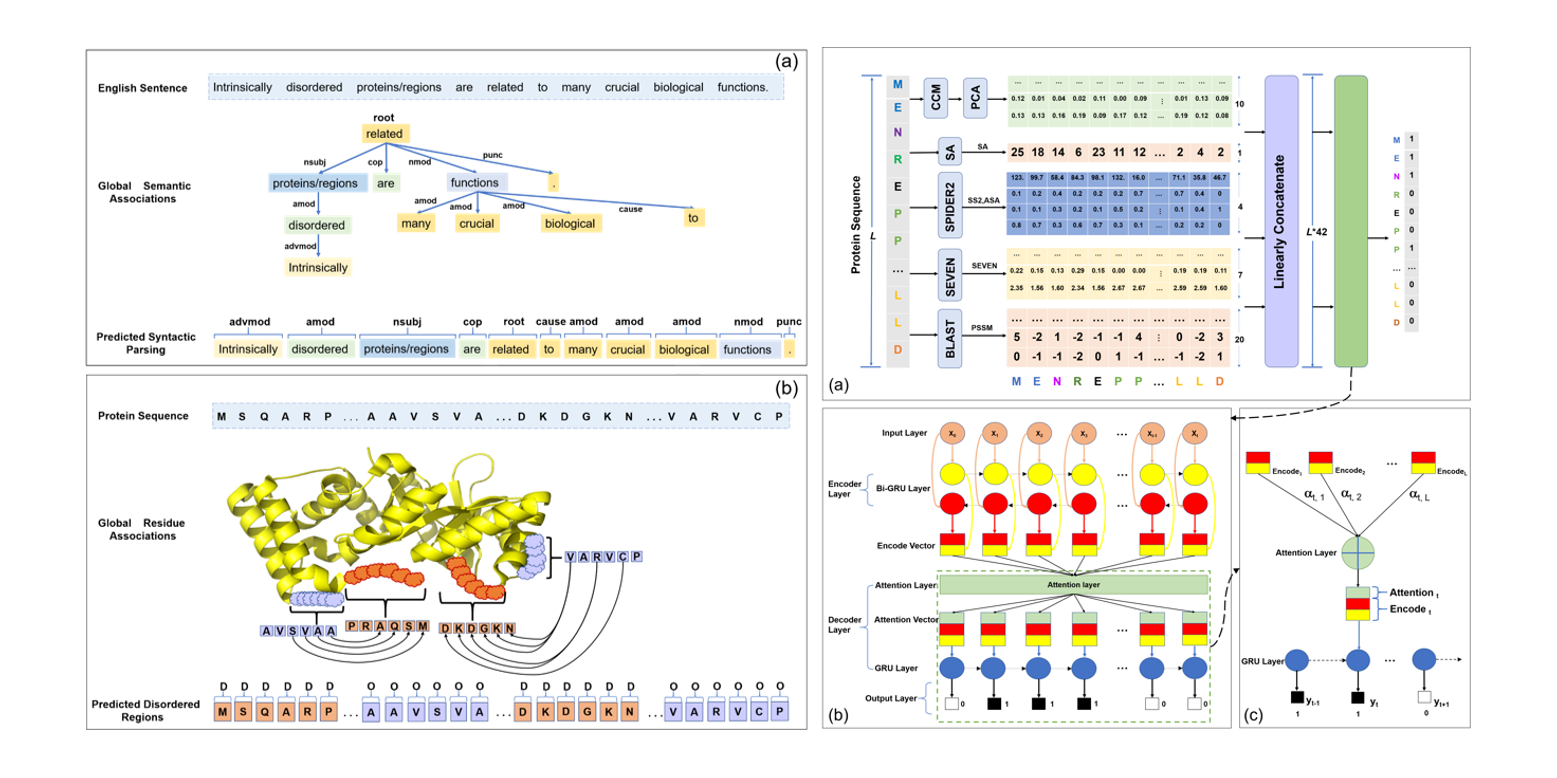
IDP-Seq2Seq
Identification of Intrinsically Disordered Proteins and Regions based on Sequence to Sequence Learning
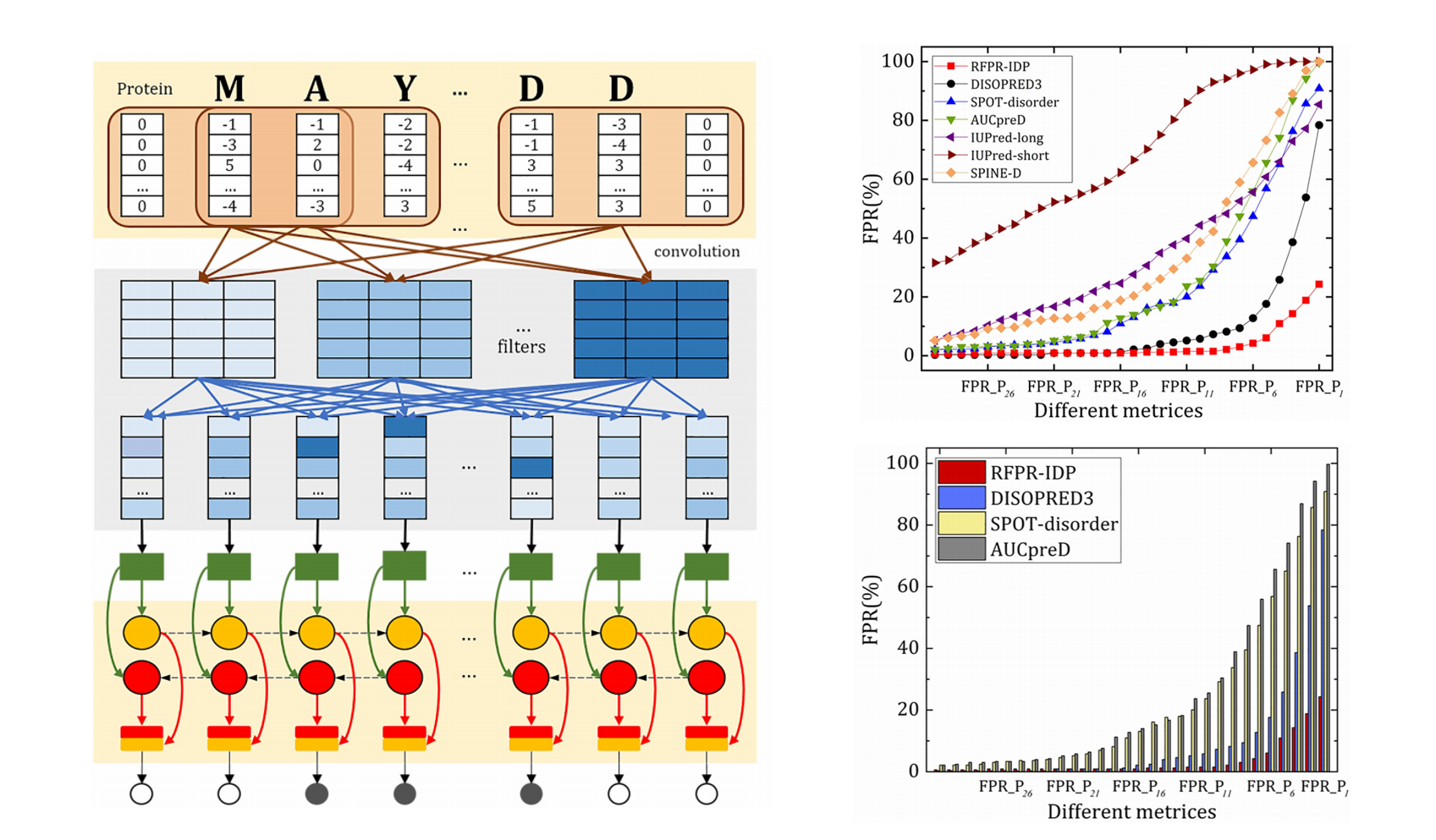
RFPR-IDP
Reduce the false positive rates for intrinsically disordered protein and region prediction by incorporating ordered proteins
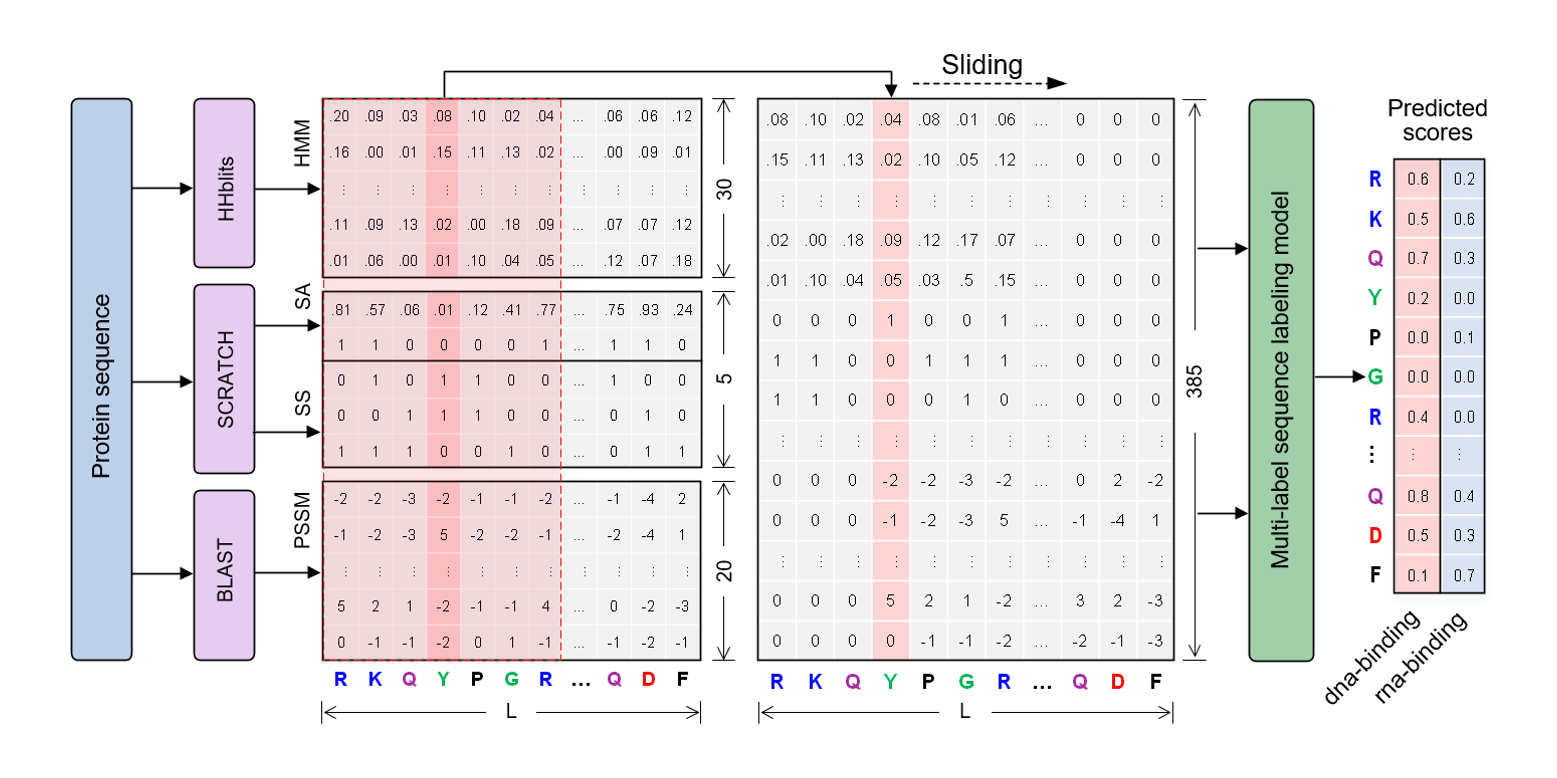
NCBRPred
identifying nucleic acid binding residues in proteins based on multi-label sequence labeling model
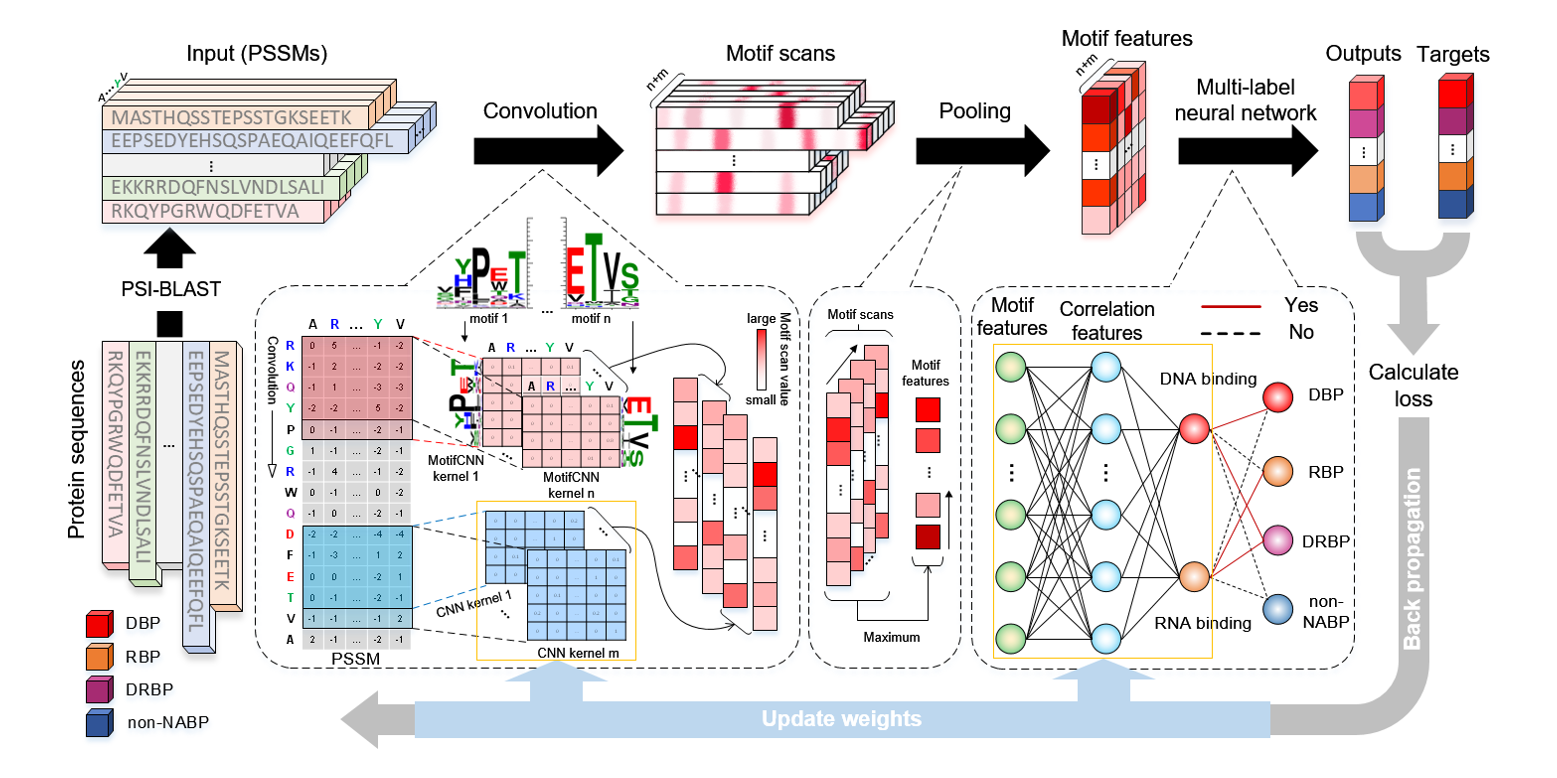
iDRBP_MMC
identifying DNA-binding proteins and RNA-binding proteins based on multi-label learning model and motif-based ...
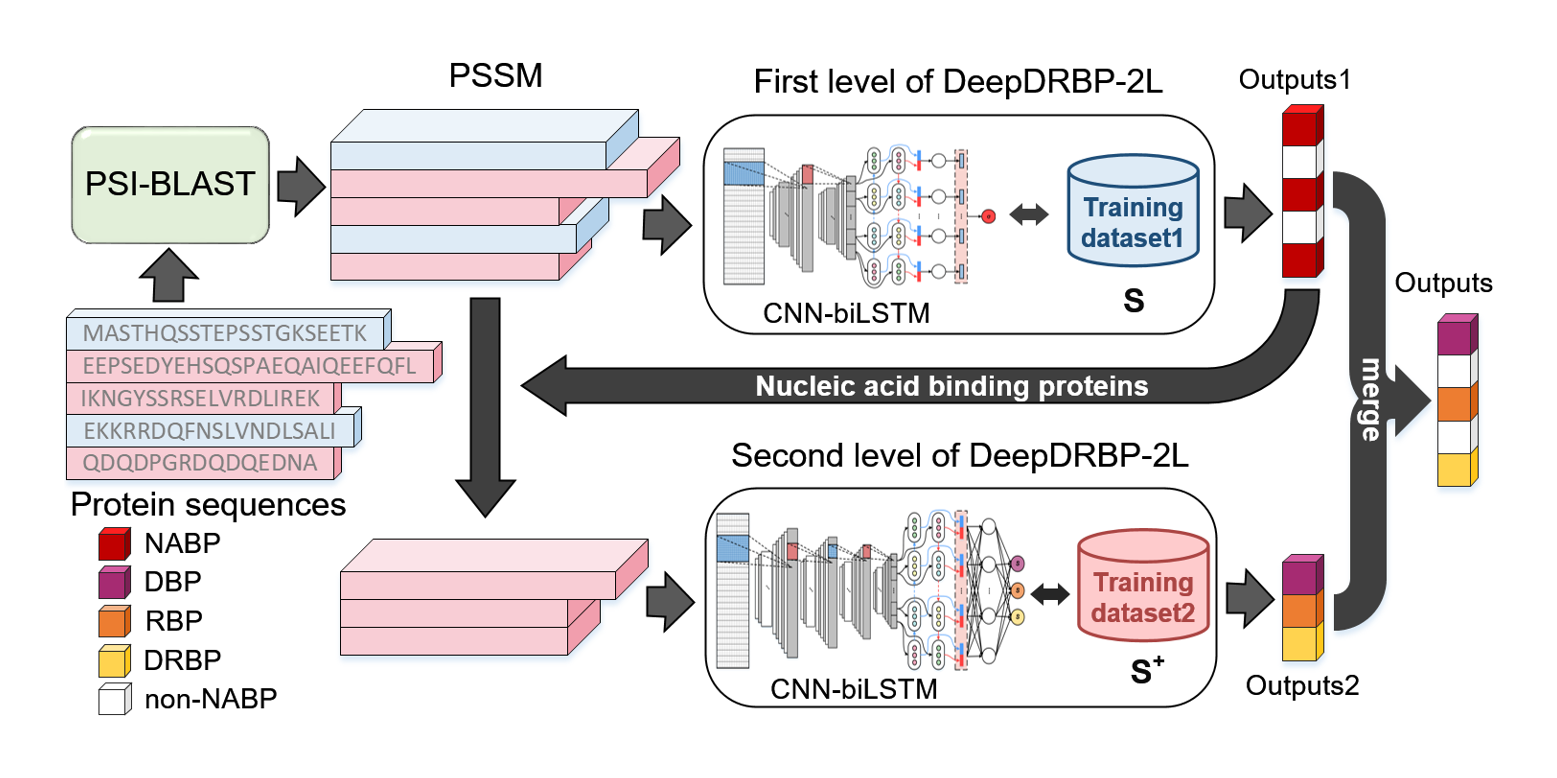
DeepDRBP-2L
a new genome annotation predictor for identifying DNA-binding proteins and RNA-binding proteins using Convolutional ...
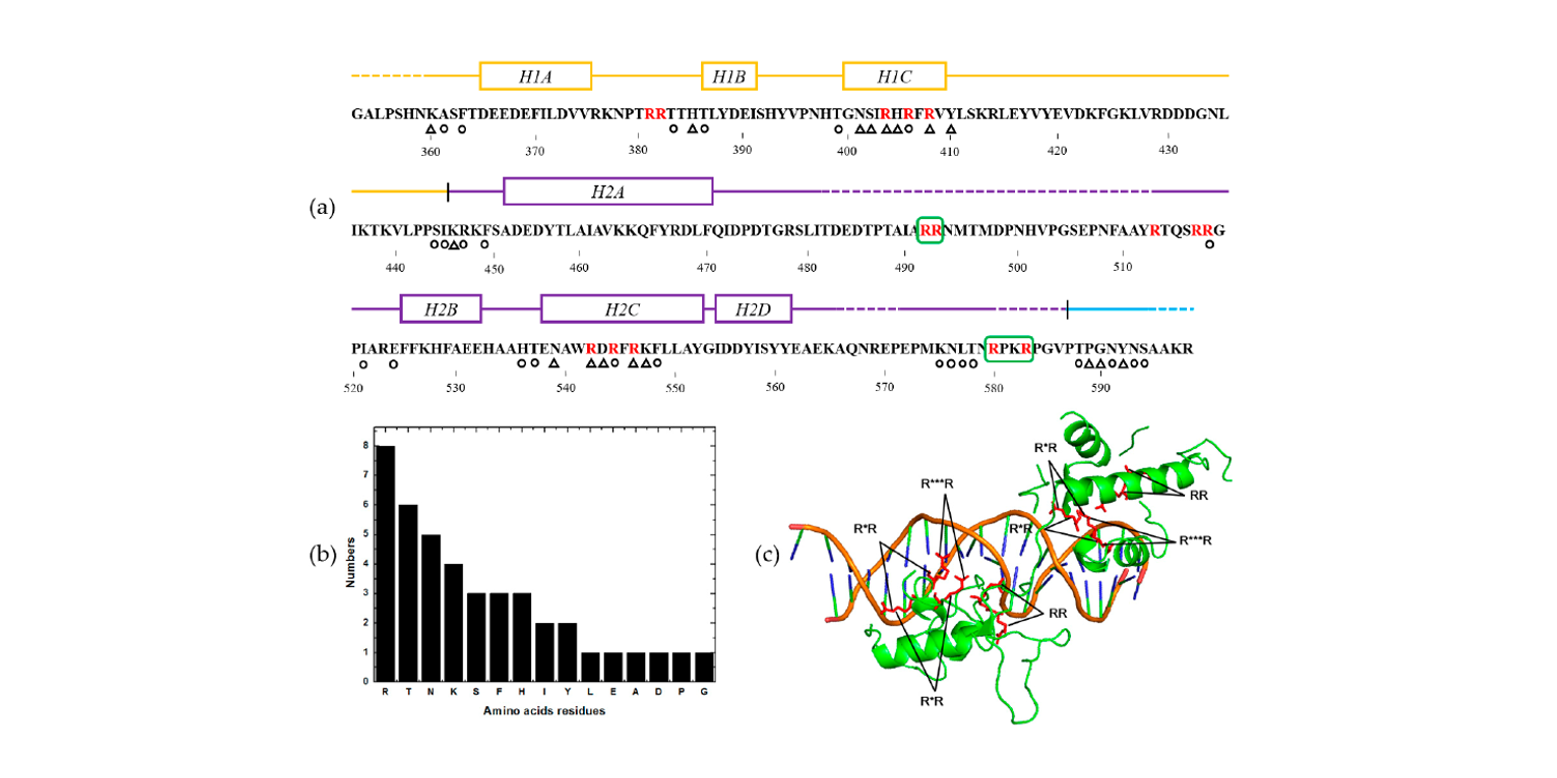
PSFM-DBT
identifying DNA-binding proteins by combing position specific frequency matrix and distance-bigram transformation
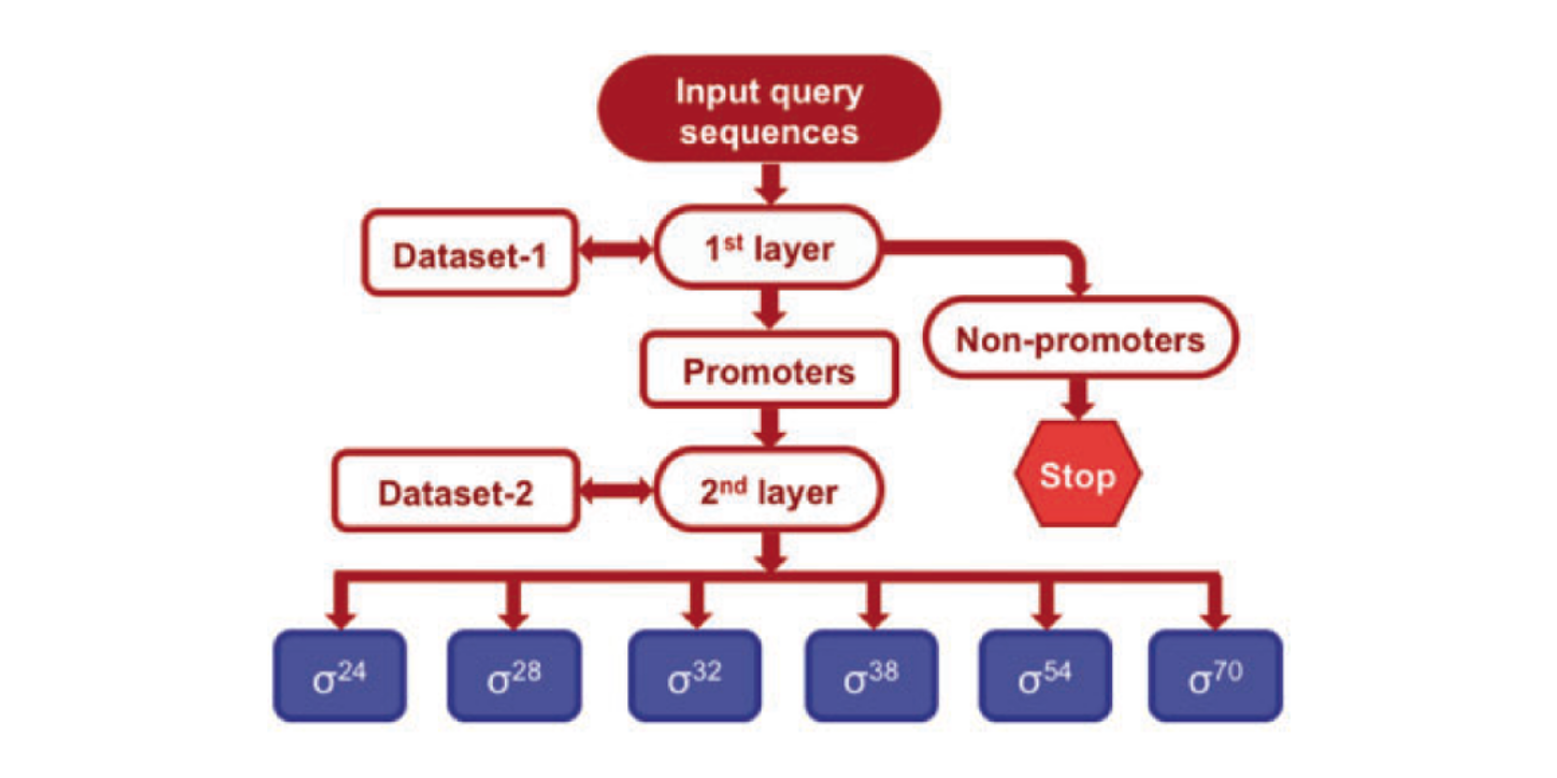
iPromoter-2L
a two-layer predictor for identifying promoters and their types by multi-window-based PseKNC
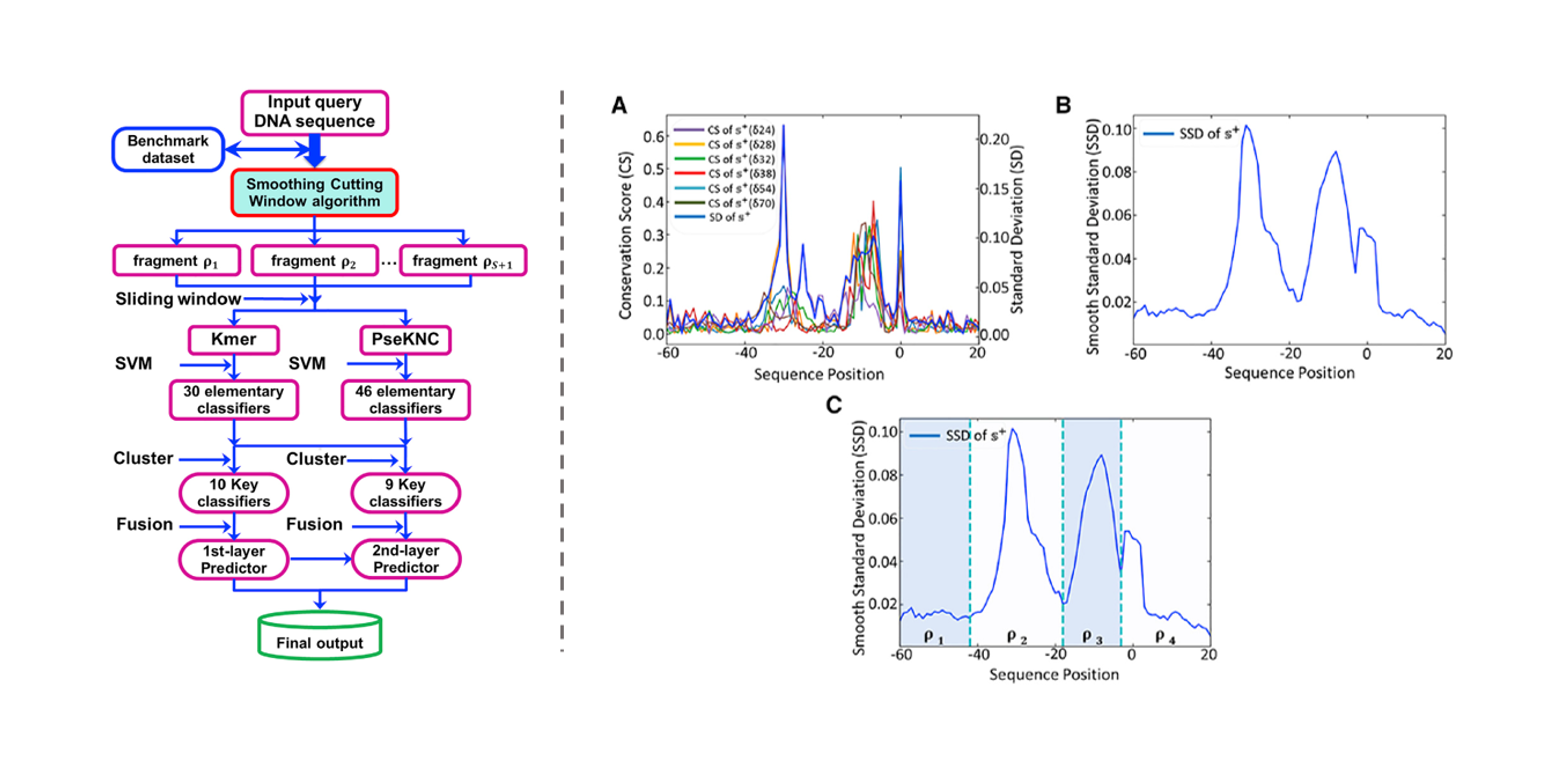
iPromoter-2L2.0
a predictor for identifying promoters and their types by combining Smoothing Cutting Window algorithm and ...
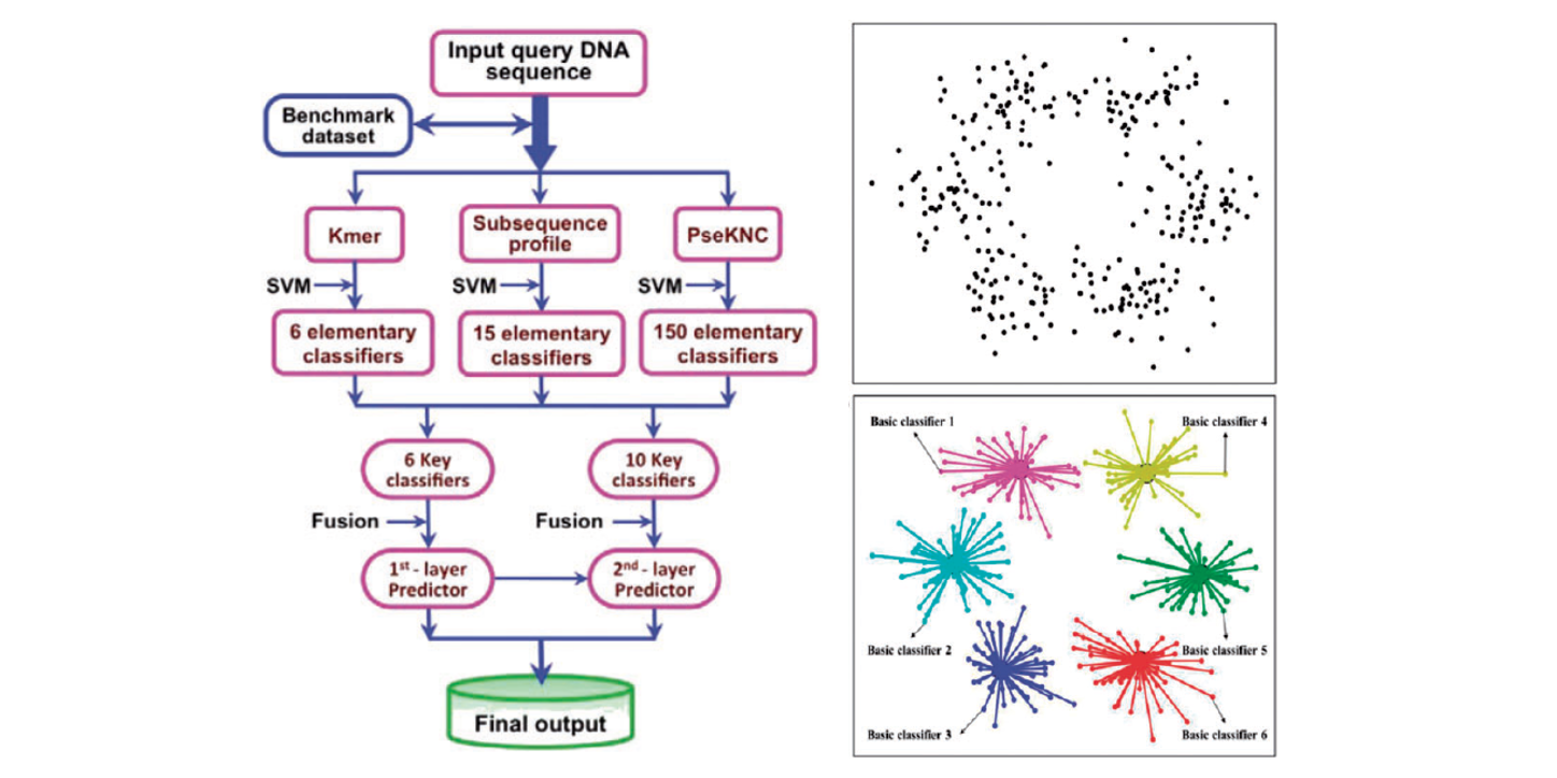
iEnhancer-EL
identifying enhancers and their strength with ensemble learning approach
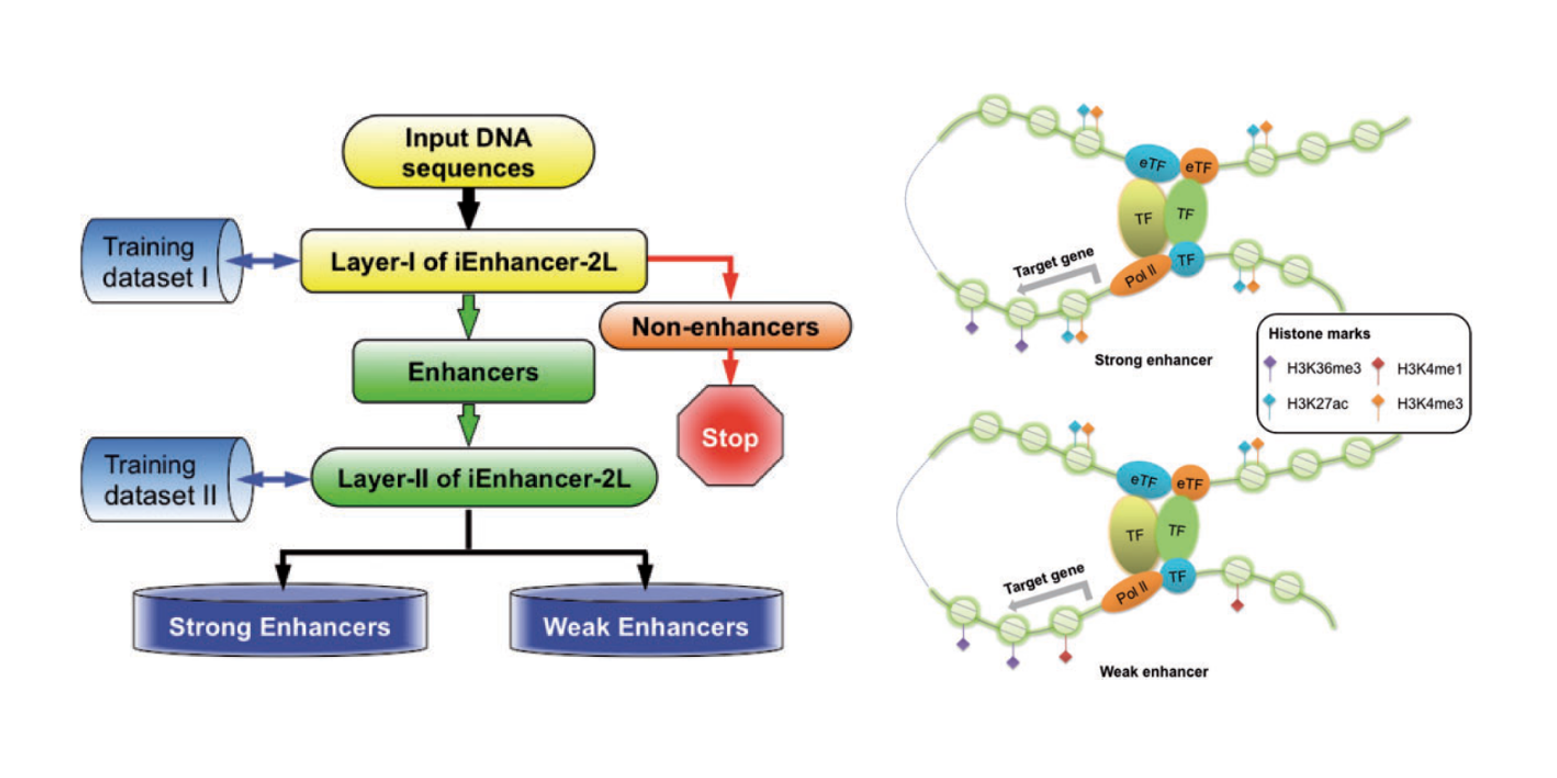
iEnhancer-2L
a two-layer predictor for identifying enhancers and their strength by pseudo k-tuple nucleotide composition
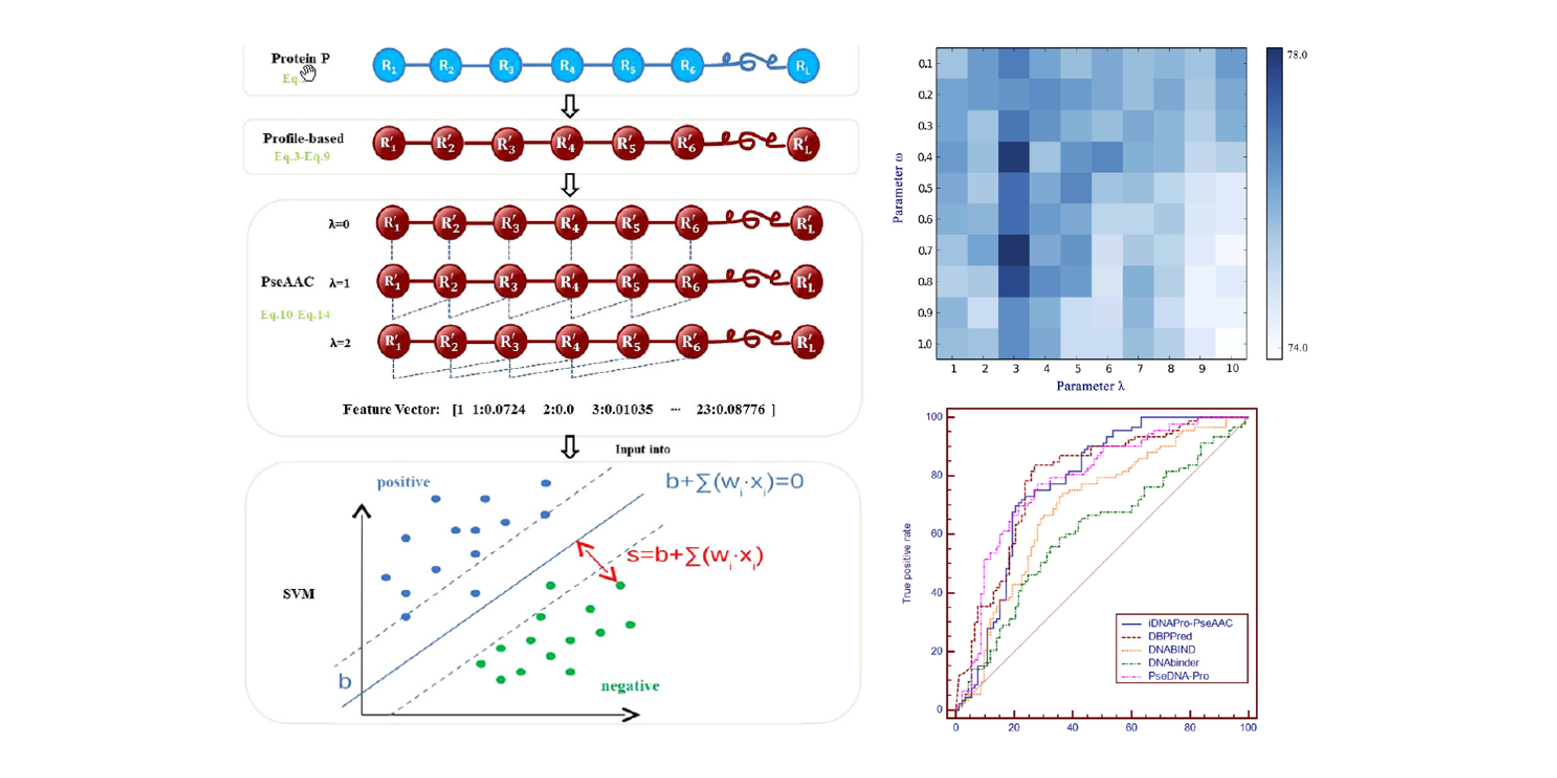
iDNAPro-PseAAC
DNA binding protein identification by combining pseudo amino acid composition and profile-based protein representation
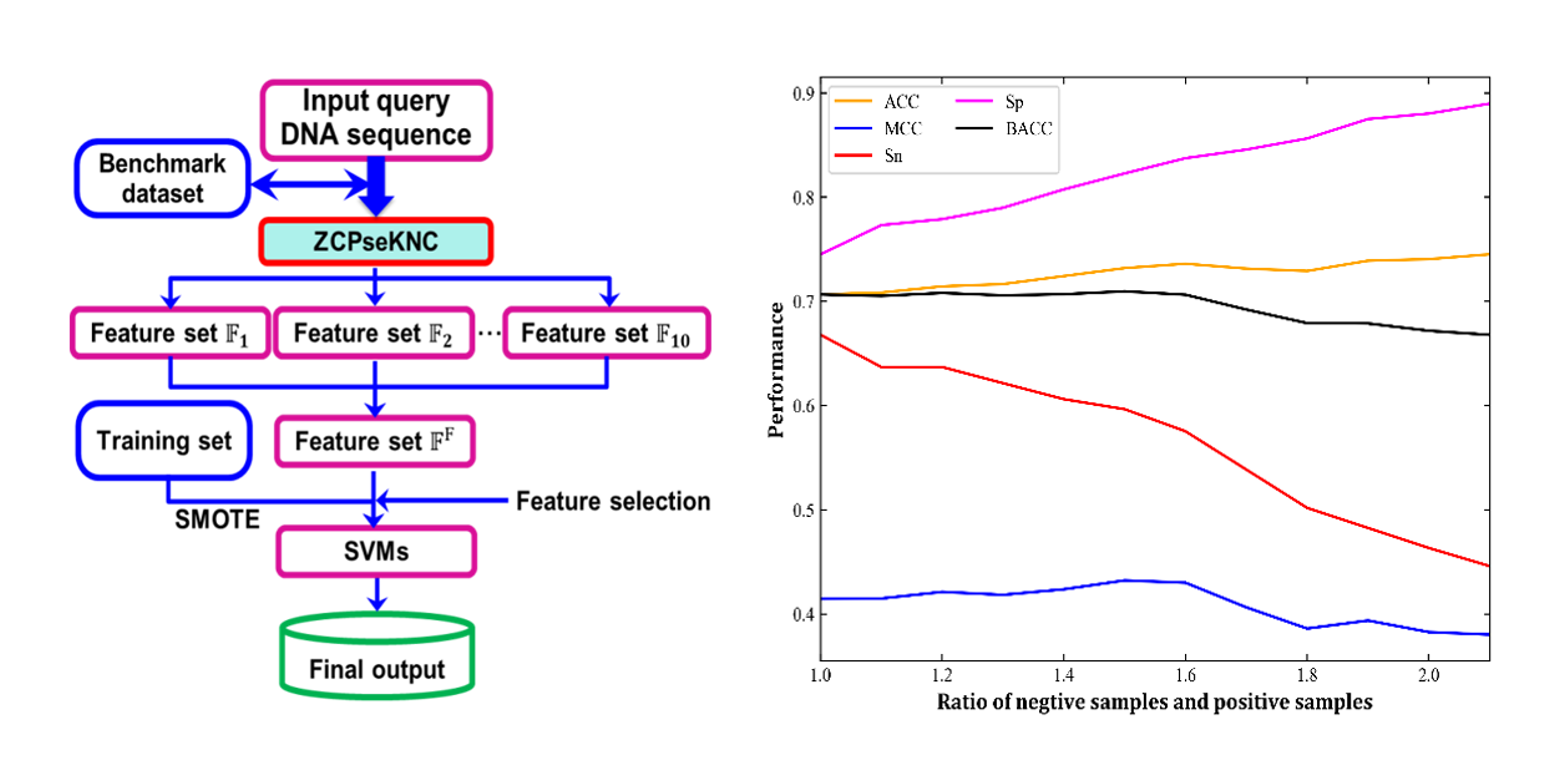
iEsGene-ZCPseKNC
identify eseential genes based on Z curve pseudo k-tuple nucleotide composition
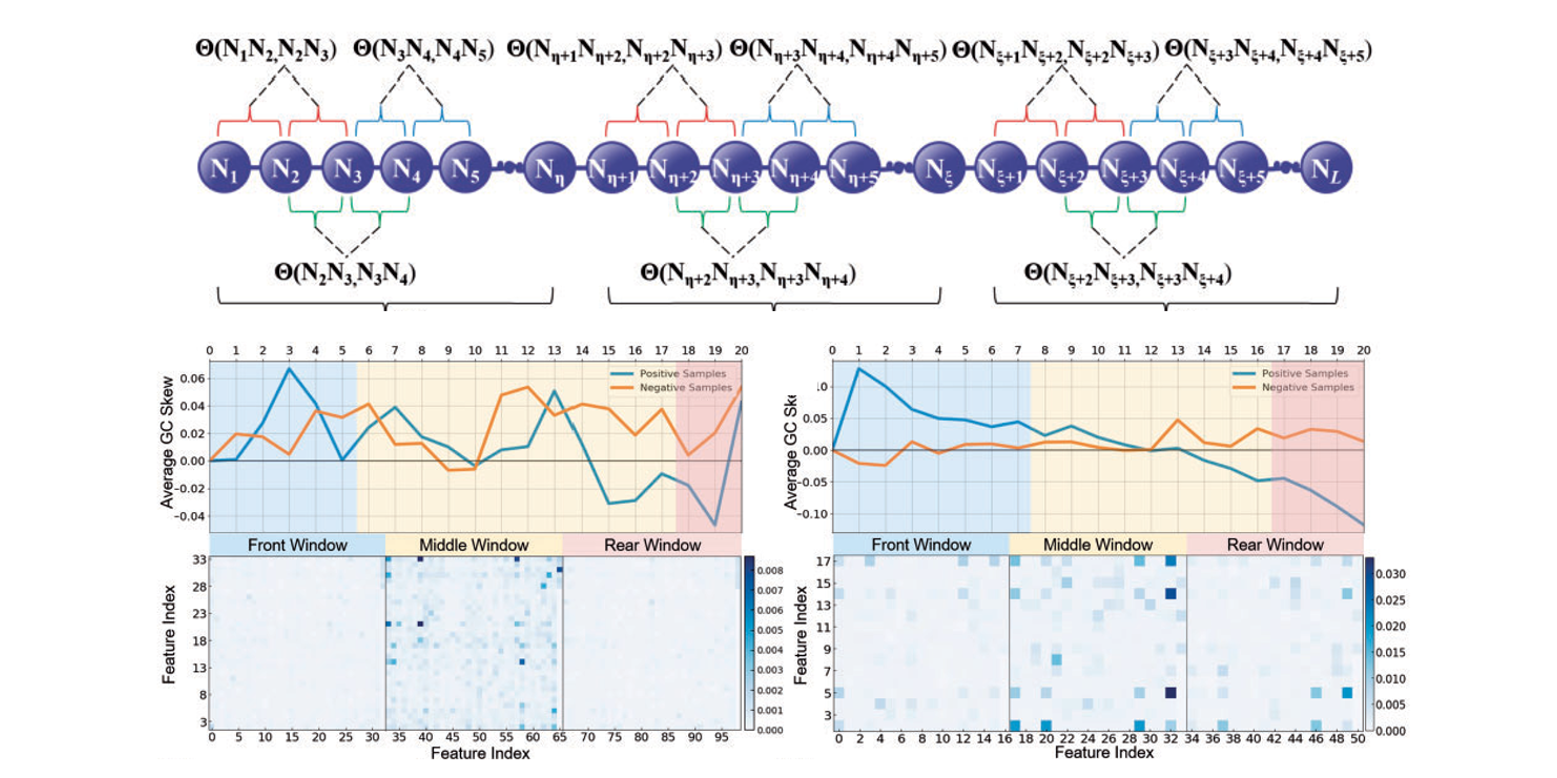
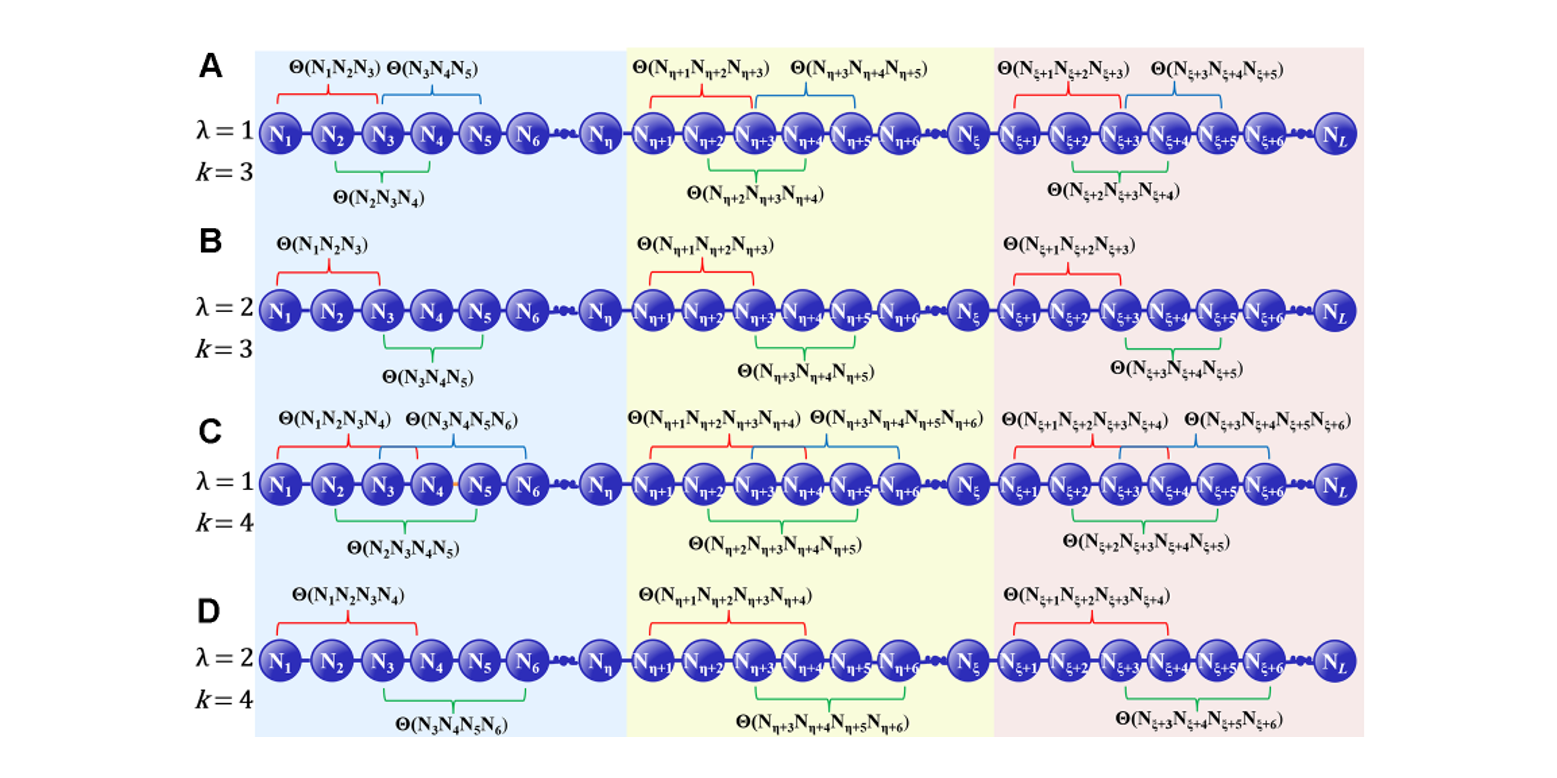
iRO-PsekGCC
identify DNA replication origins based on Pseudo k-tuple GC Composition
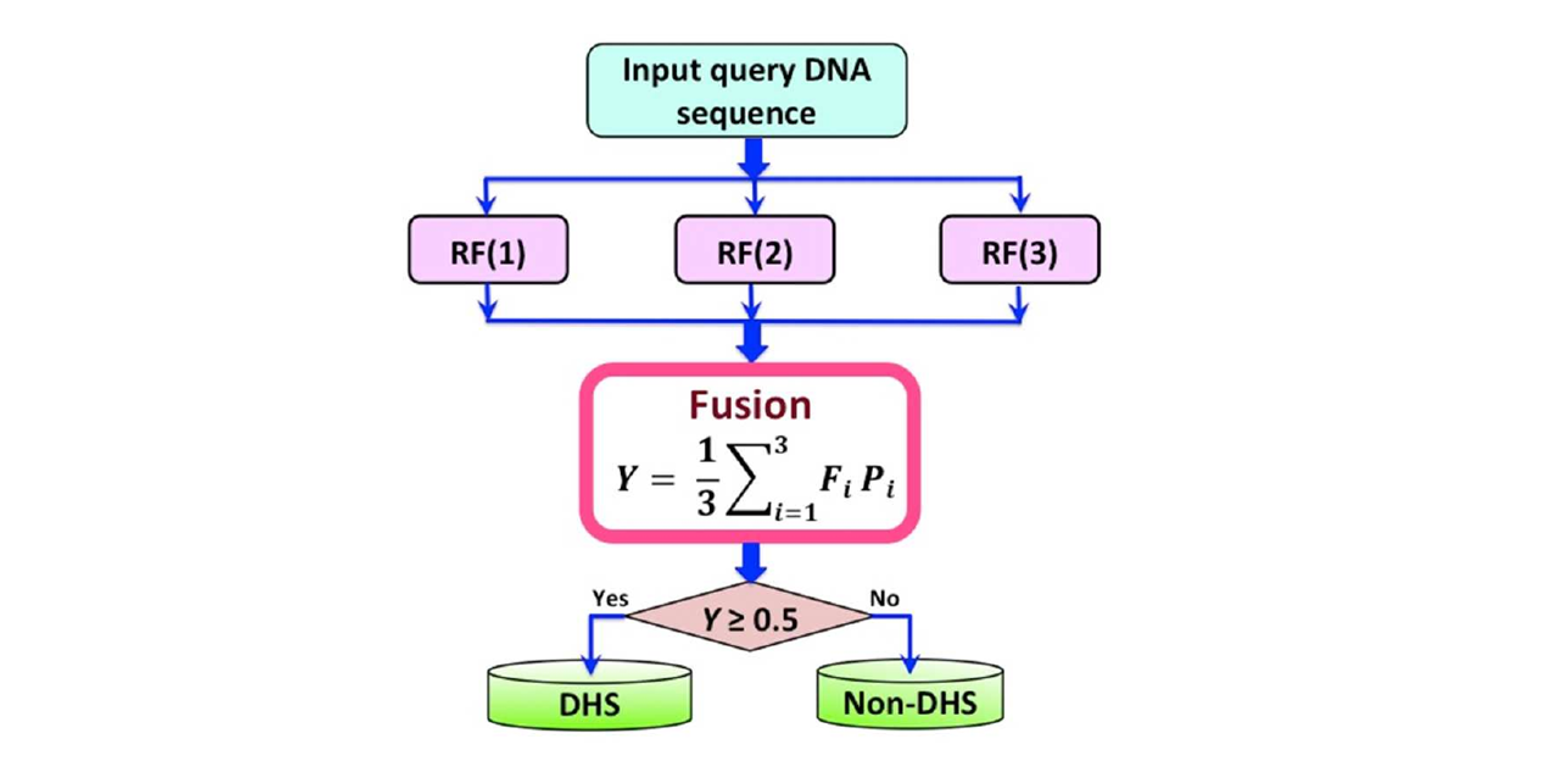
iDHS-EL
Identifying DNase I hypersensitive sites by fusing three different modes of pseudo nucleotide composition into an ensemble ...
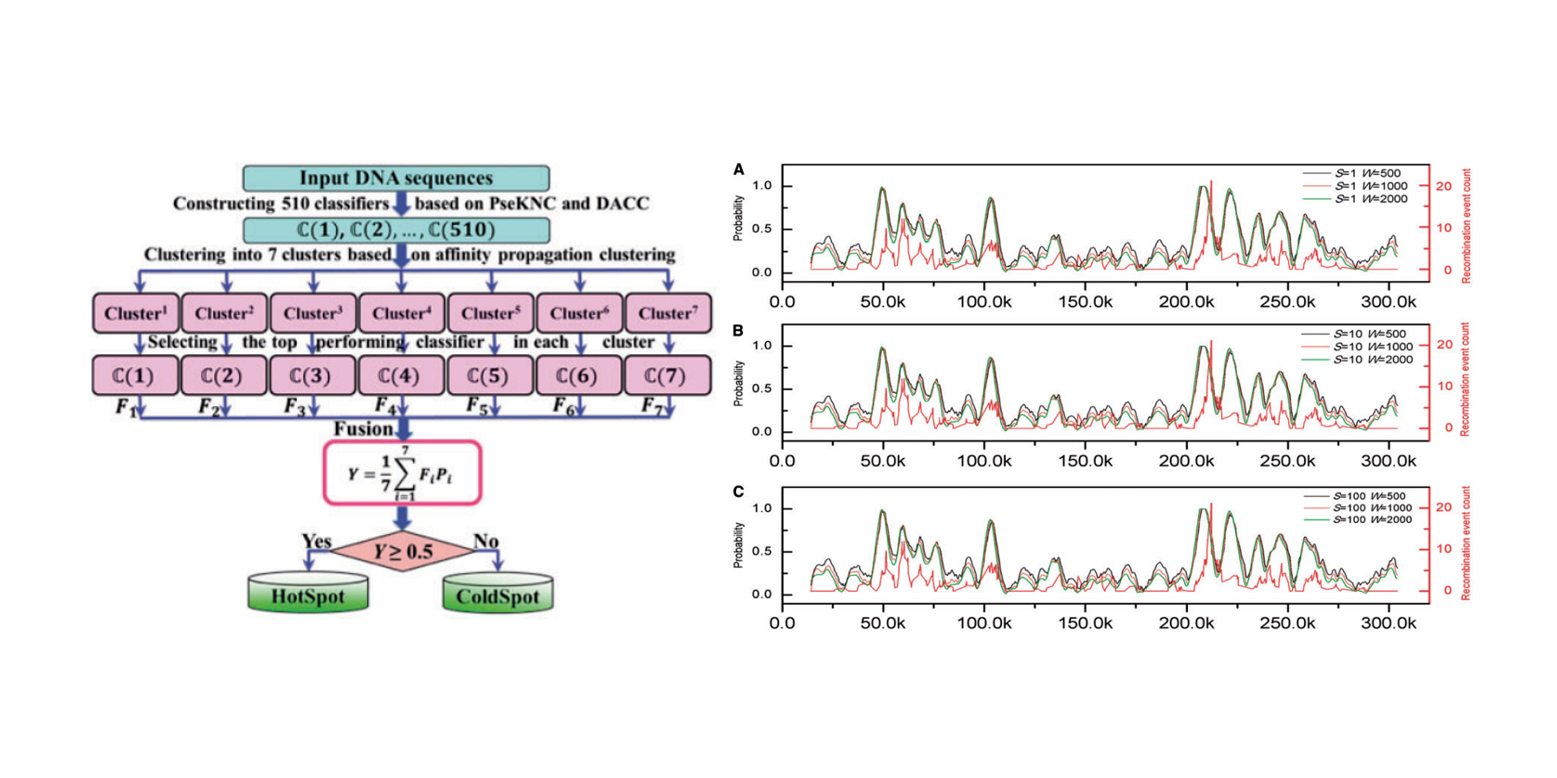
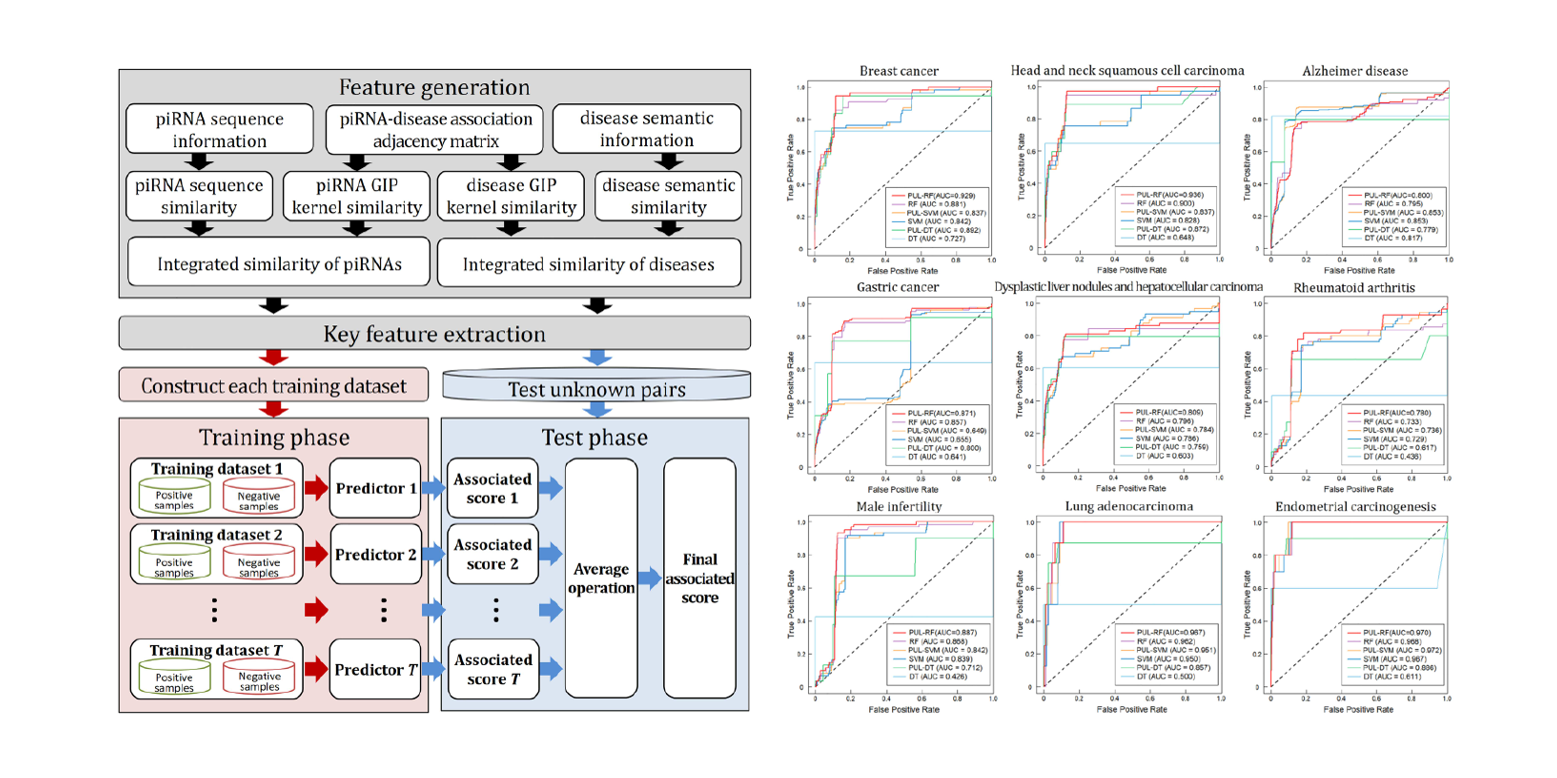
iPiDi-PUL
identifying Piwi-interacting RNA-disease associations based on Positive Unlabeled Learning
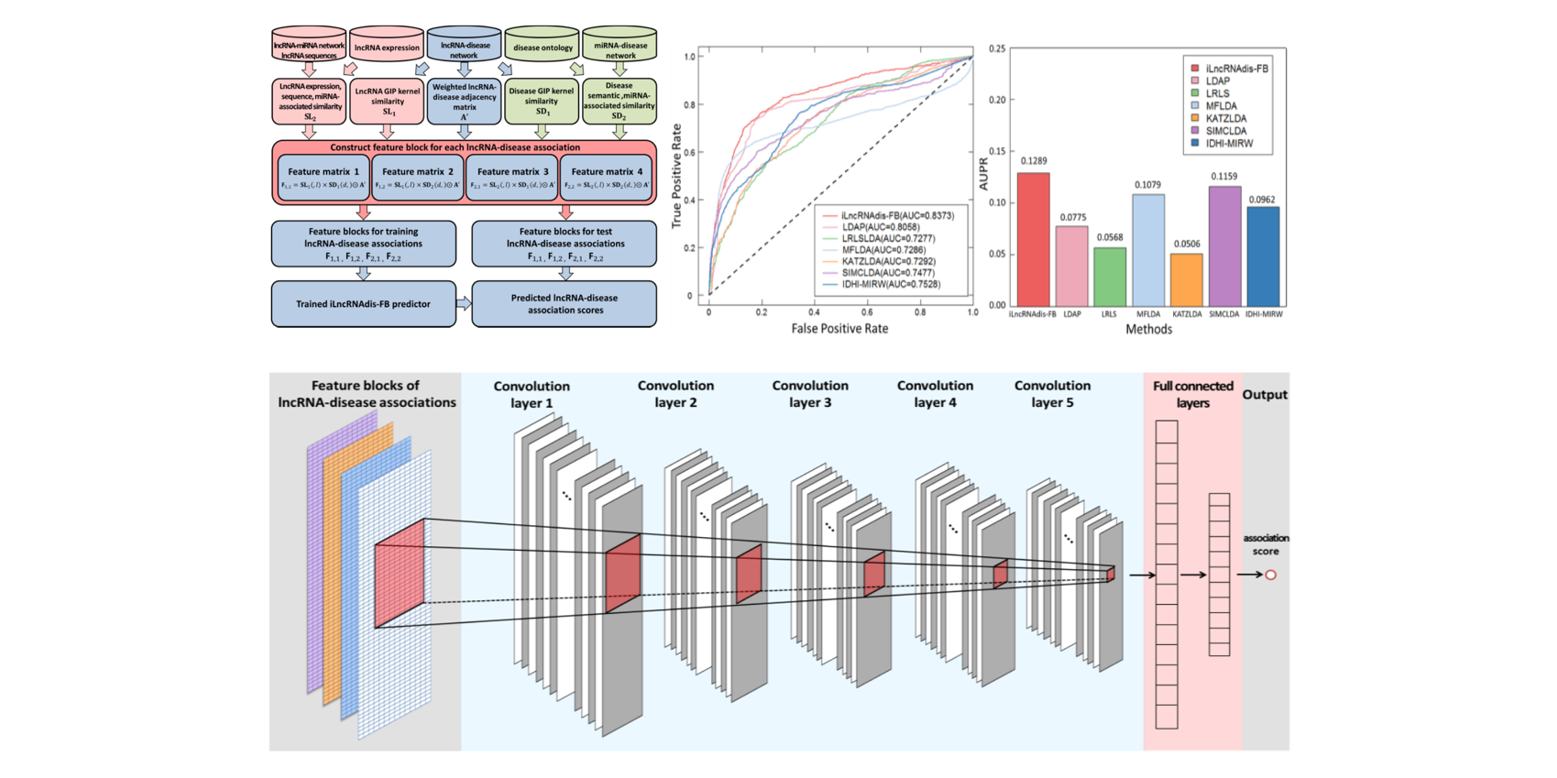
iLncRNAdis-FB
a new predictor for identifying lncRNA-disease associations by fusing biological feature blocks through deep neural network
miRNA-deKmer
identification of microRNA precursor with the degenerate K-tuple or Kmer strategy
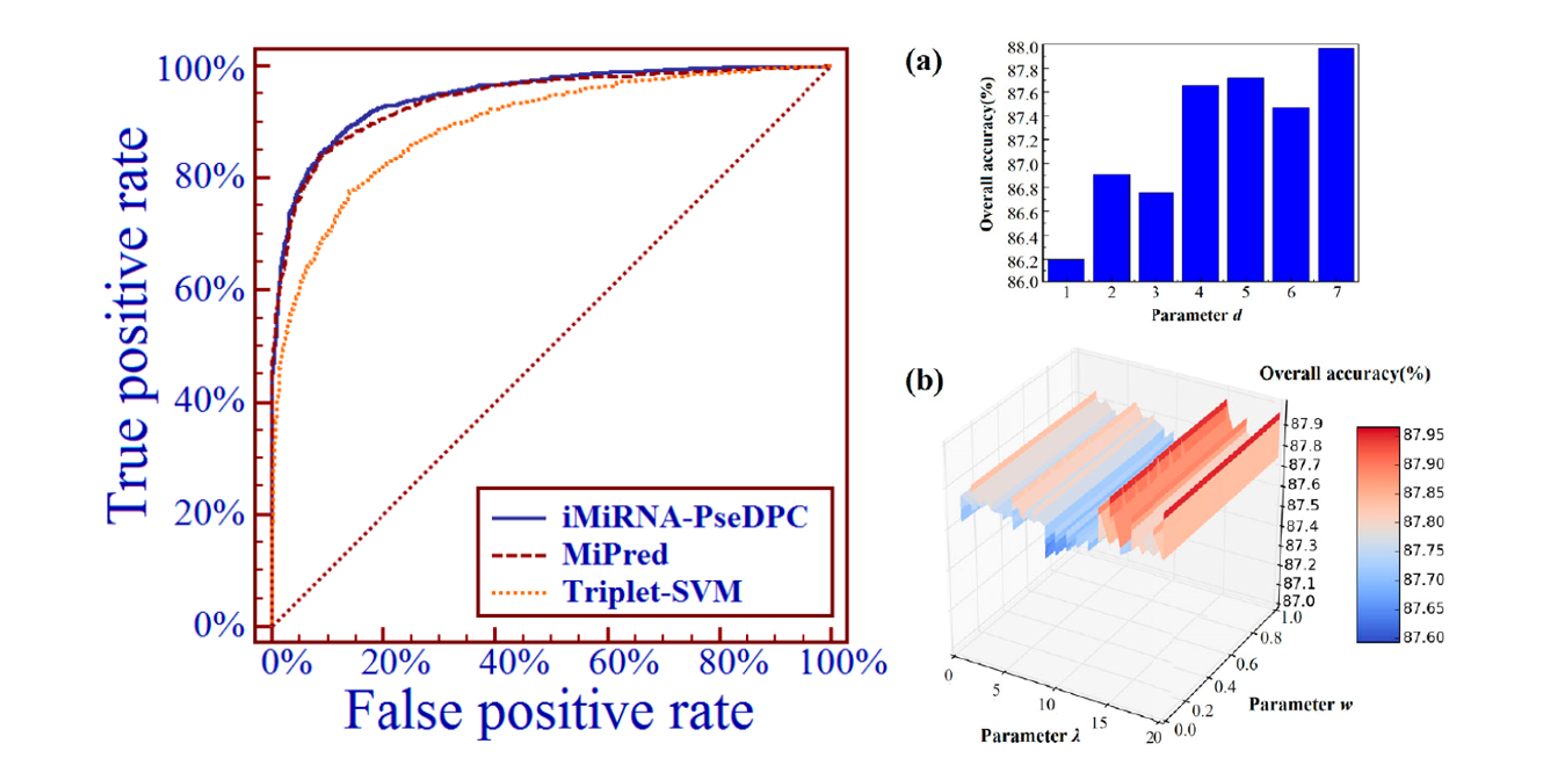
iMiRNA-PseDPC
microRNA precursor identification with a pseudo distance-pair composition approach
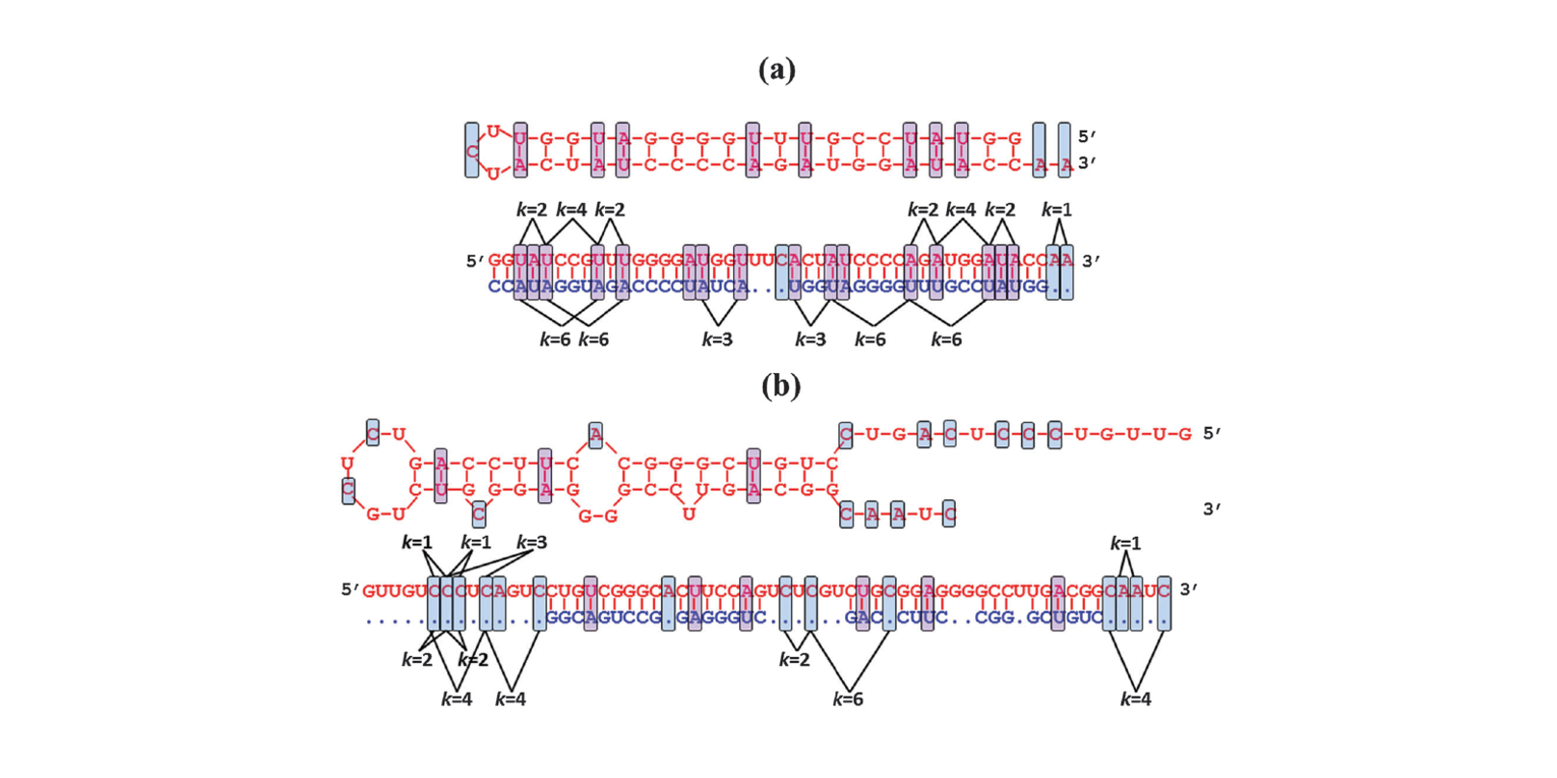
miRNA-dis
microRNA precursor identification based on distance structure status pairs
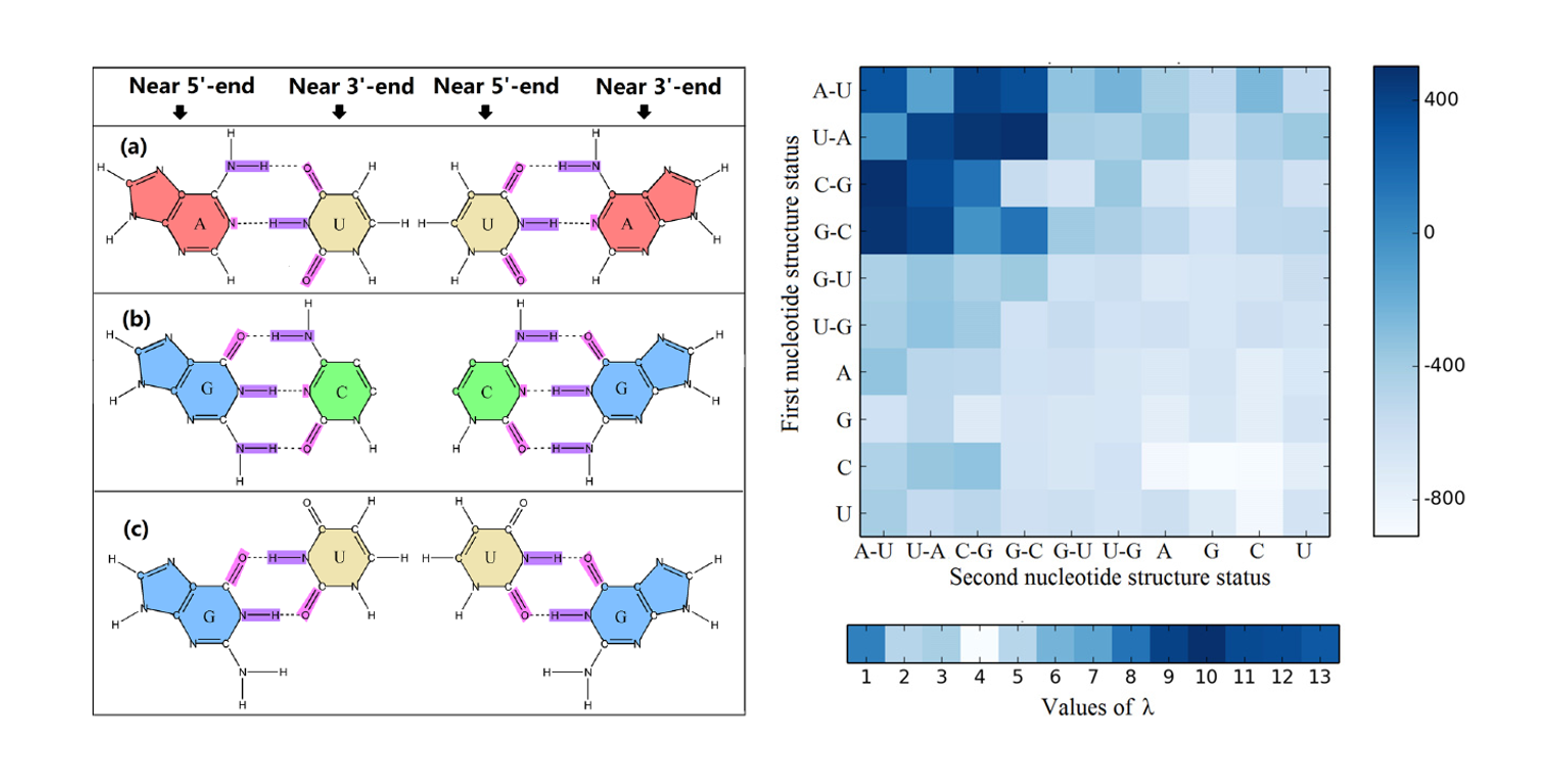
iMcRNA
identification of the real microRNA precursors with a pseudo structure status composition approach
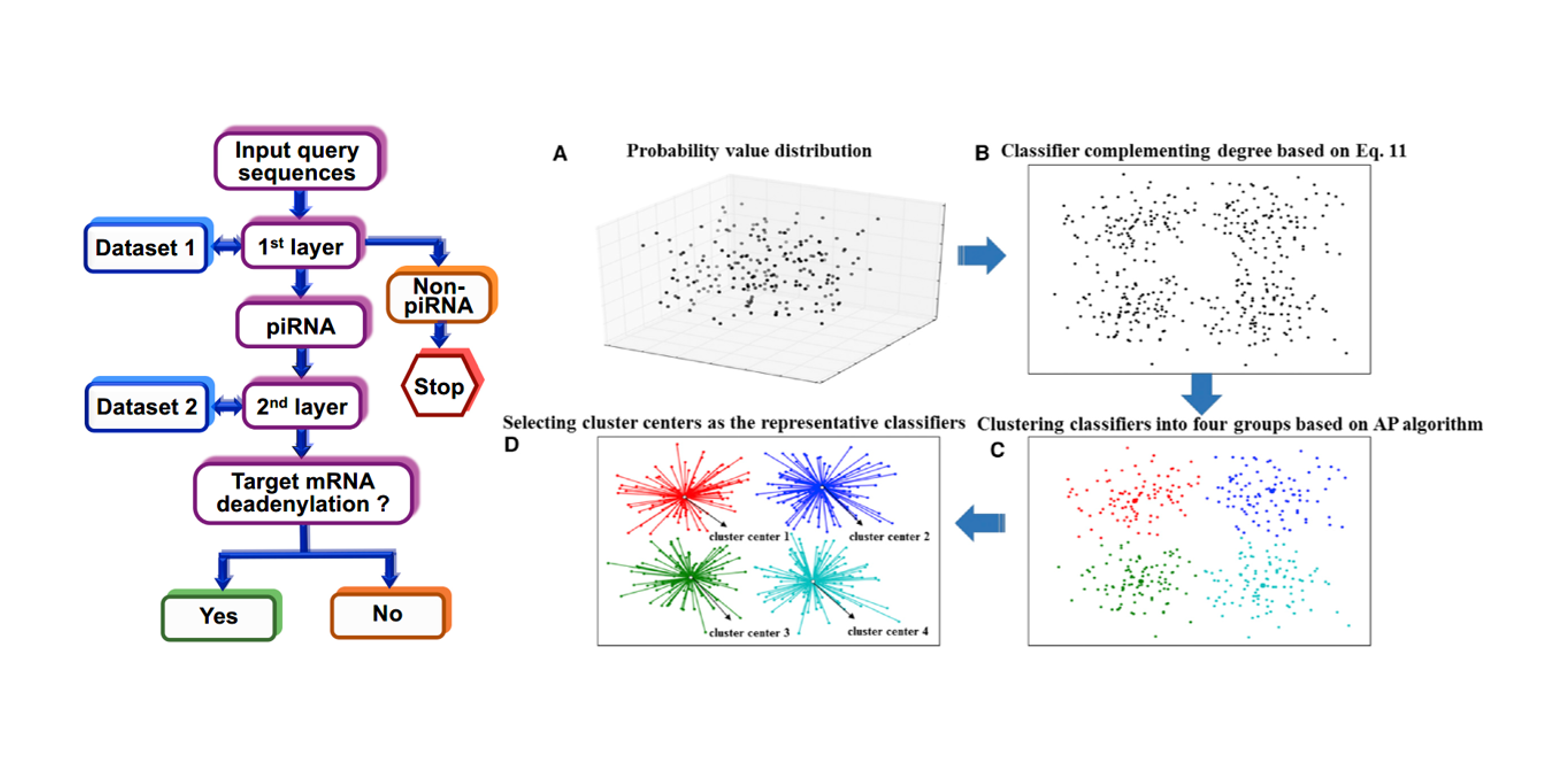
2L-piRNA
a two-layer ensemble classifier identifying piwi-interacting RNAs and their function
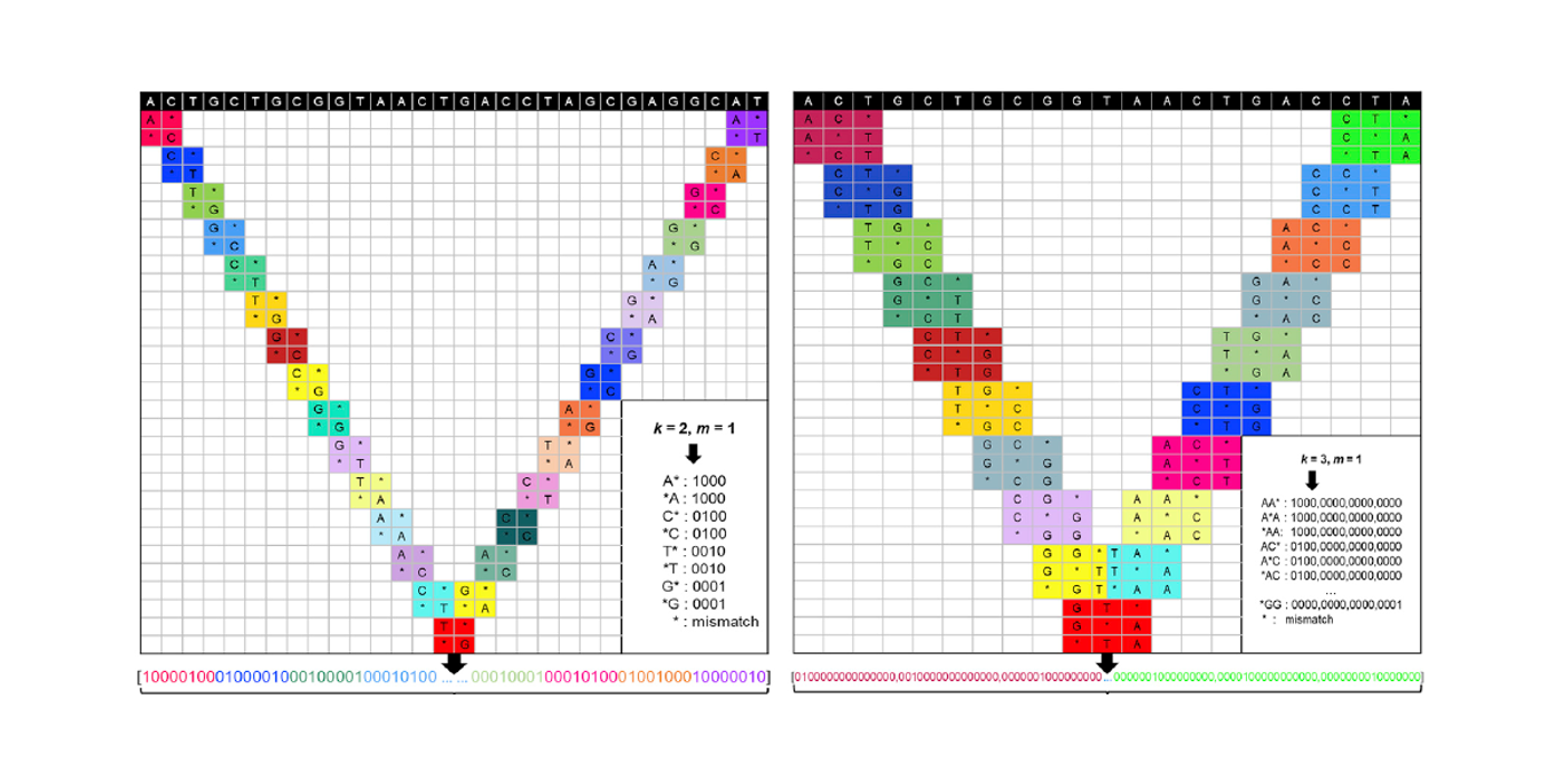
sgRNA-PSM
predict sgRNAs on-target activity based on Position Specific Mismatch
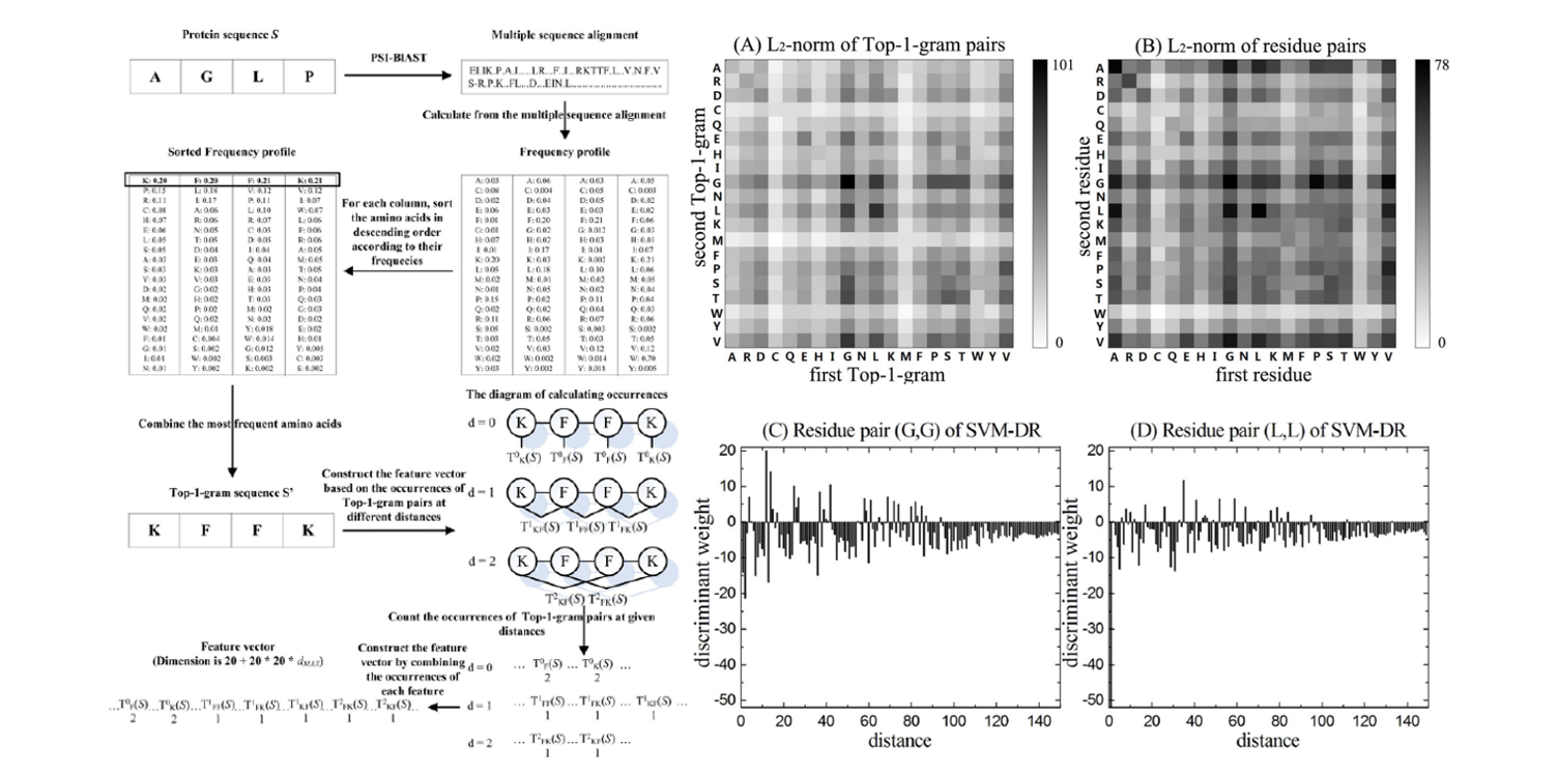
DistanceSVM
Using distances between Top-n-gram and residue pairs for protein remote homology detection
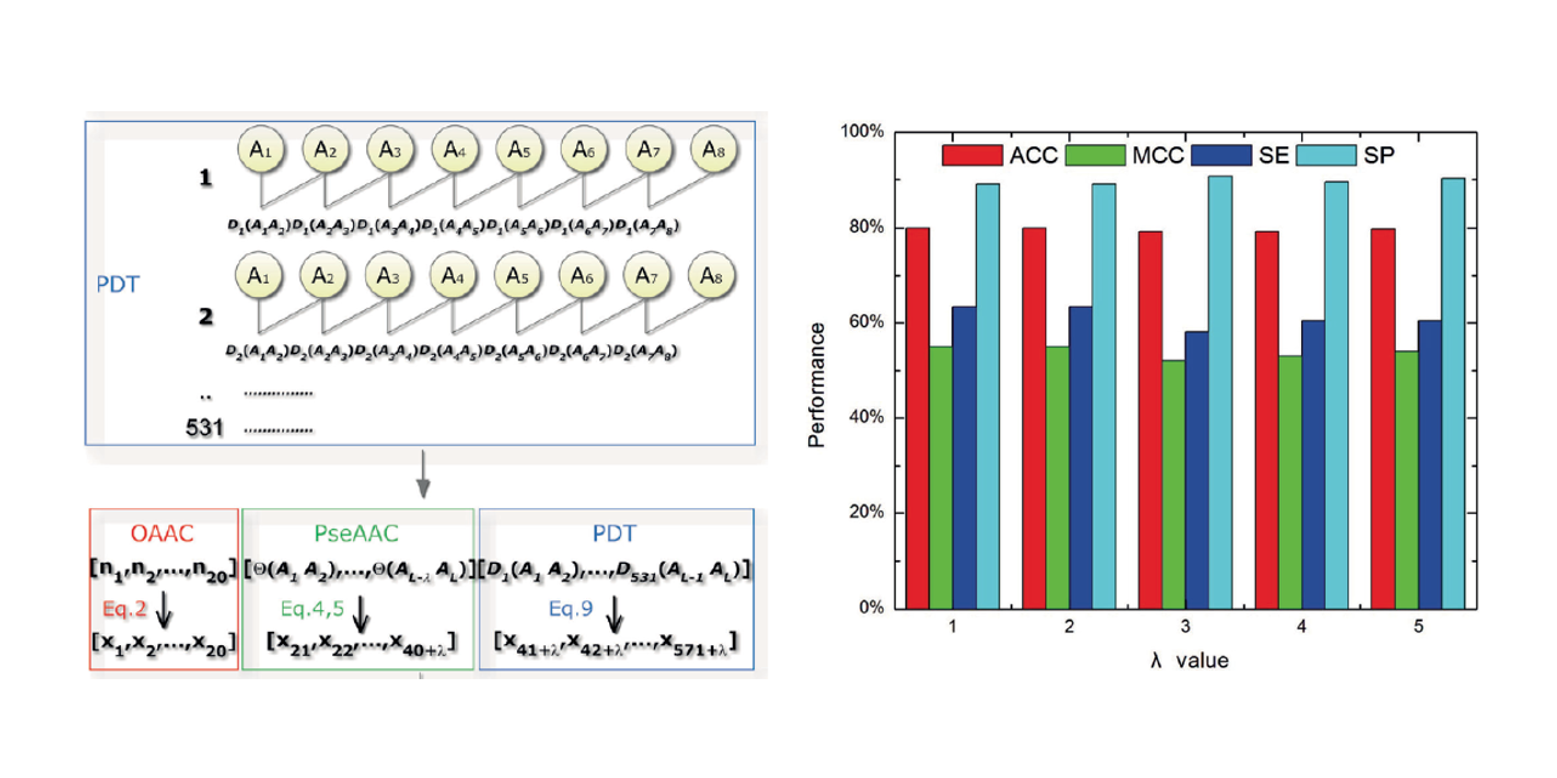
PseDNA-Pro
DNA-binding Protein Identification by Combining Chou's PseAAC and Physicochemical Distance Transformation
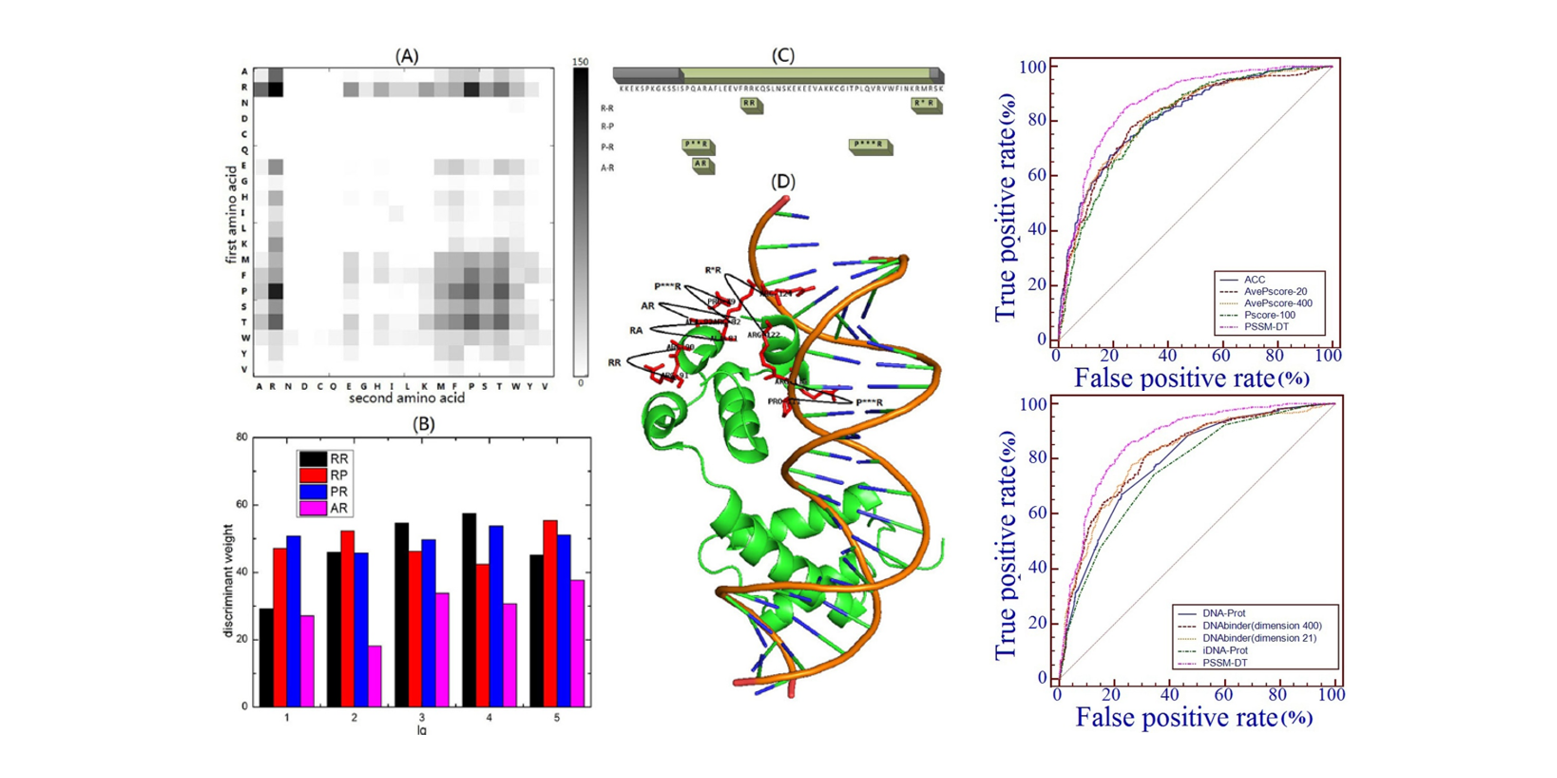
PSSM-DT
Identifying DNA-binding proteins by combining support vector machine and PSSM distance transformation
iMiRNA-SSF
Improving the Identification of MicroRNA Precursors by Combining Negative Sets with Different Distributions
iDNA-Prot|dis
identifying DNA-binding proteins by incorporating amino acid distance-pairs and reduced alphabet profile into the ...
enDNA-Prot
Identification of DNA-binding Proteins by Applying Ensemble Learning
remote
Combining evolutionary information extracted from frequency profiles with sequence-based kernels for protein remote ...
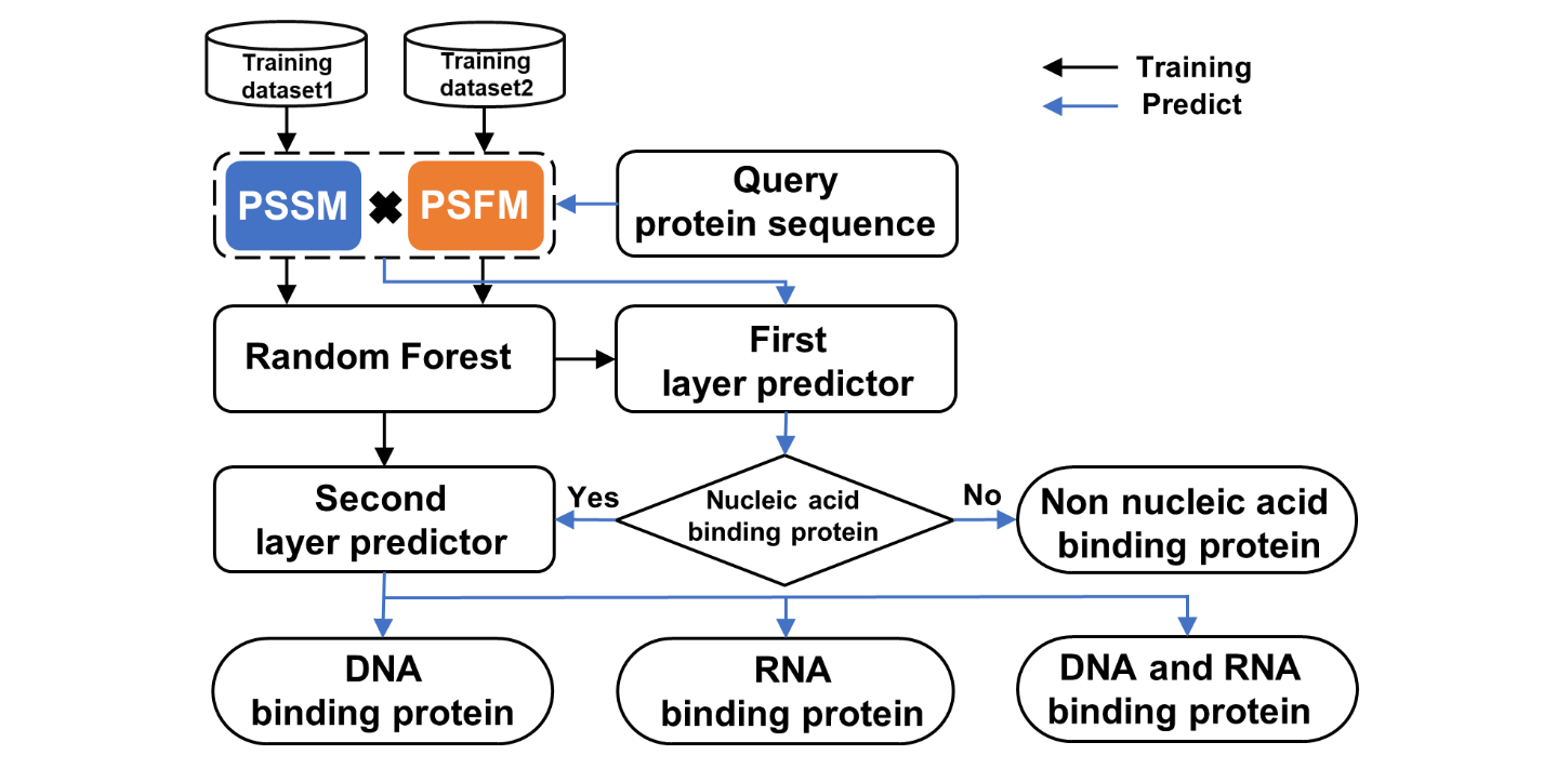
IDRBP-PPCT
Identifying nucleic acid-binding proteins based on PSSM and PSFM Cross Transformation
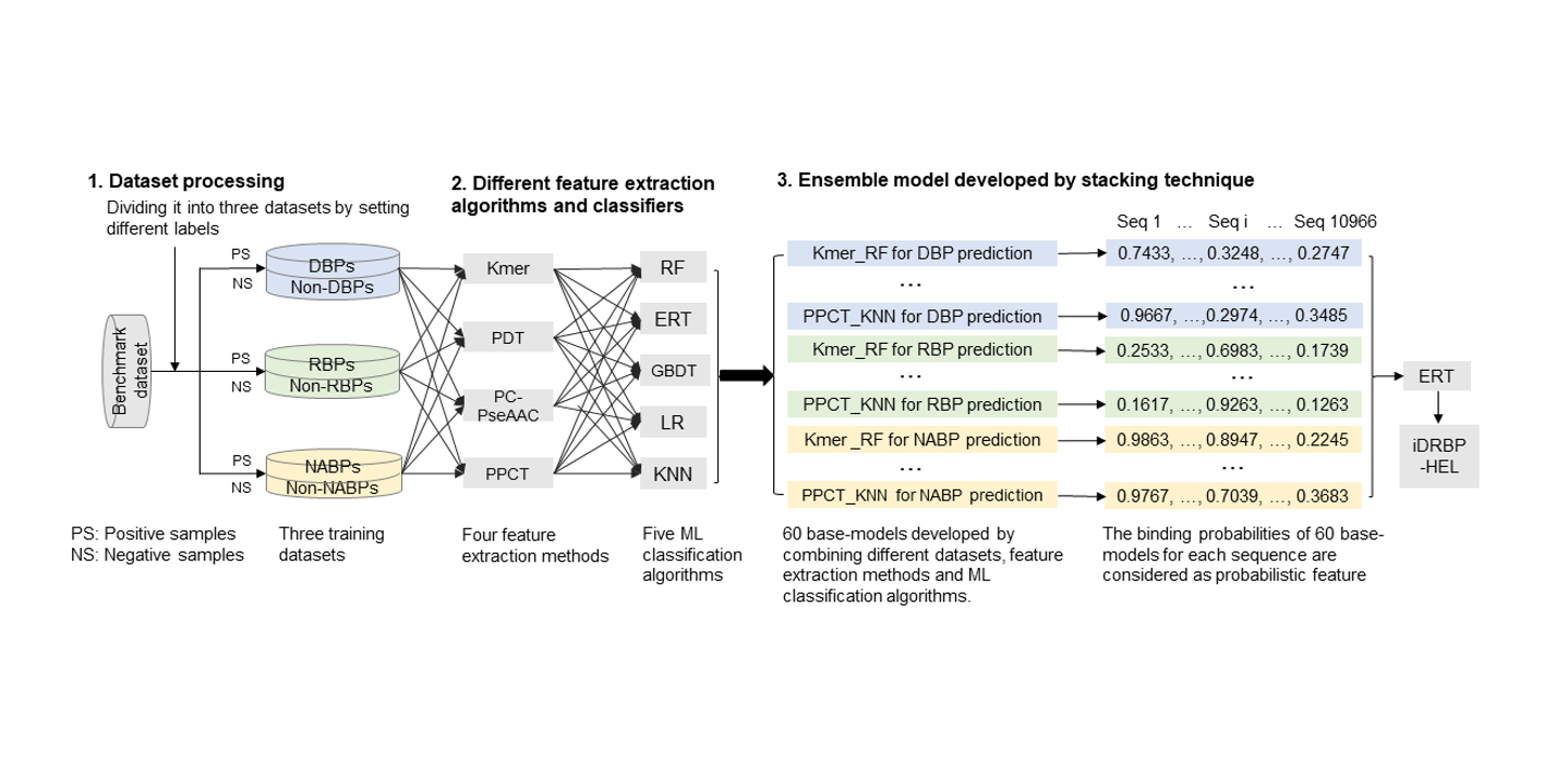
iDRBP-EL
identifying DNA-binding proteins and RNA-binding proteins based on hierarchical ensemble learning
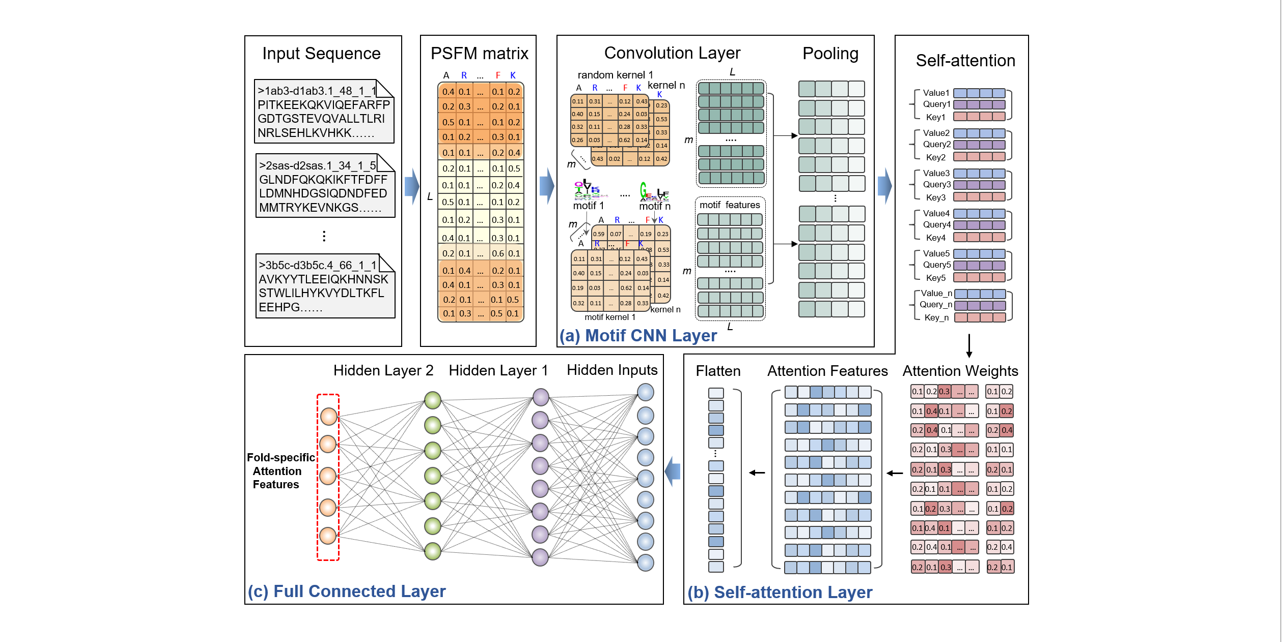
selfAT-fold
protein fold recognition based on residue-based and motif-based self-attention networks
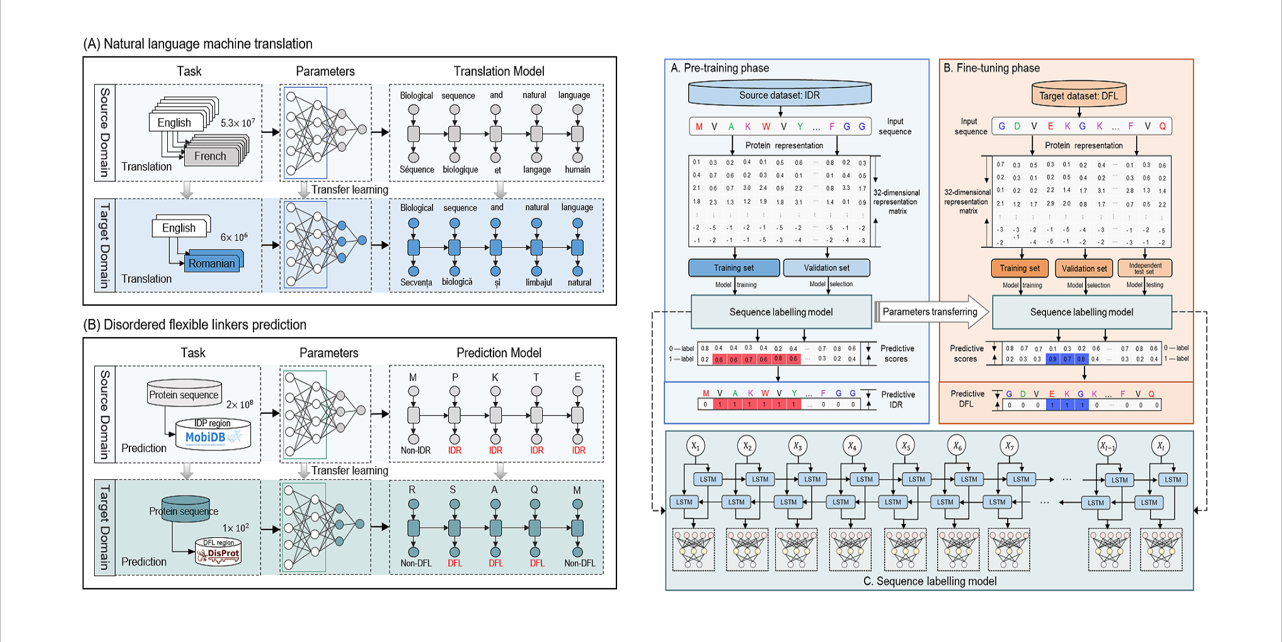
TransDFL
identification of disordered flexible linker regions in proteins by combining sequence labeling and transfer learning
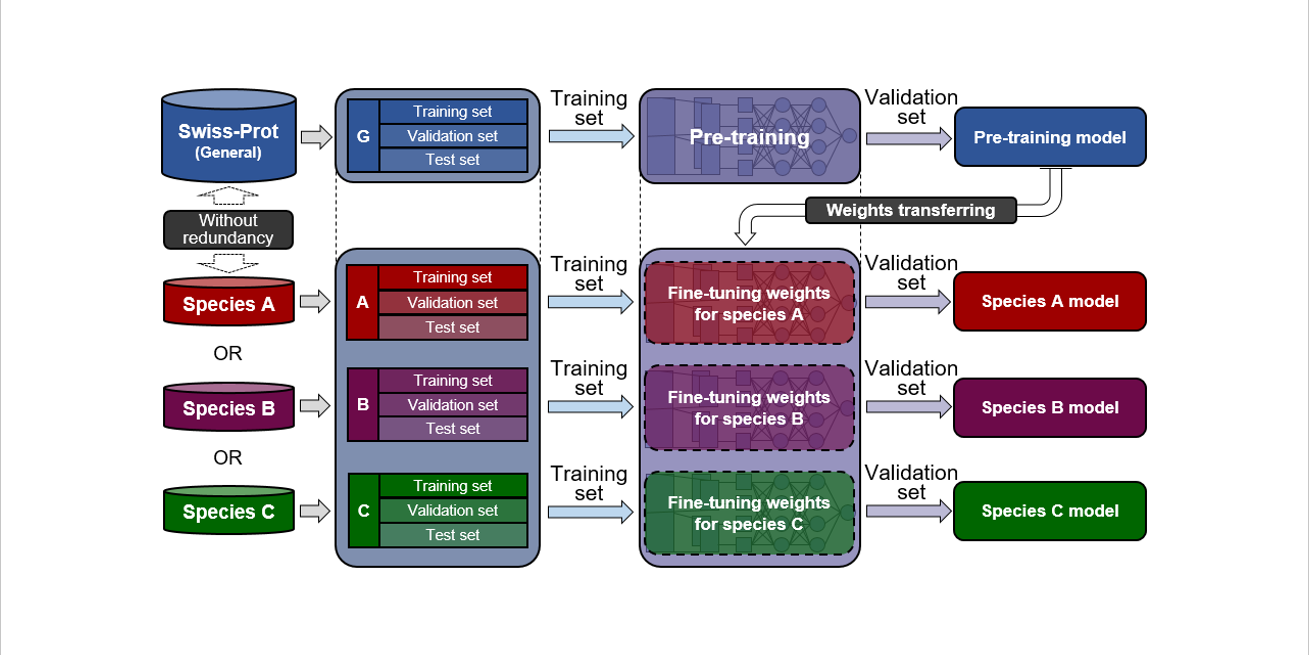
PreRBP-TL
prediction of species-specific RNA-binding proteins based on transfer learning
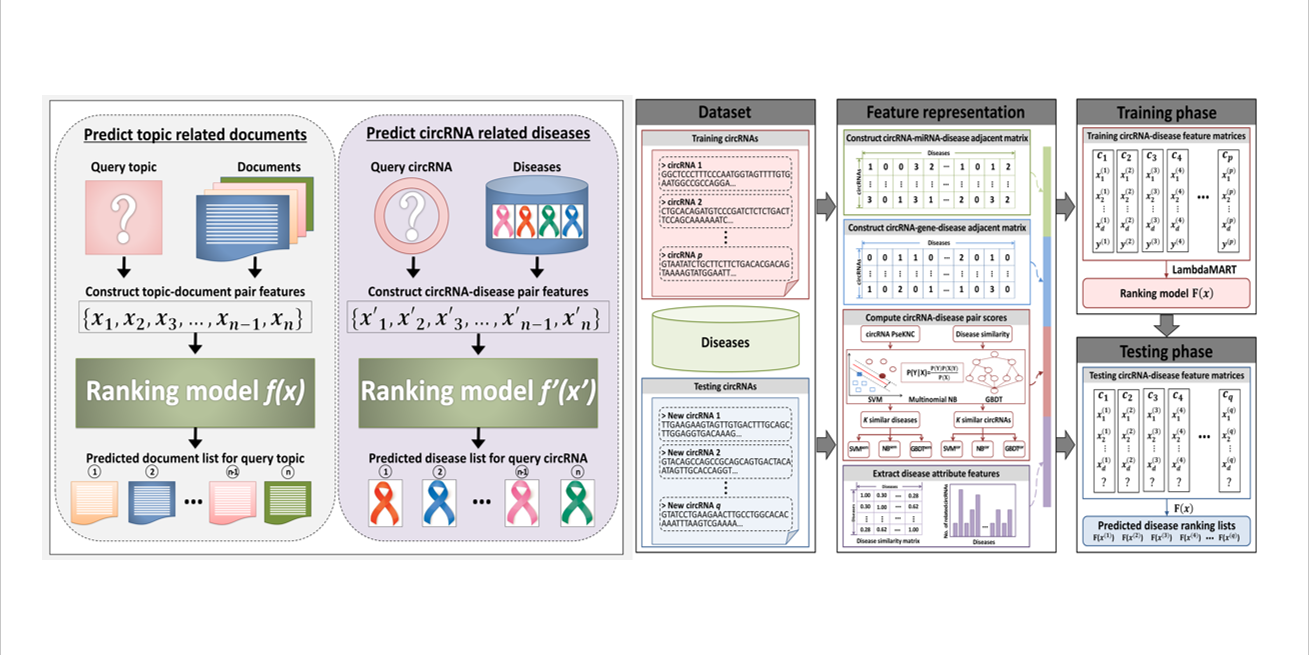
iCircDA-LTR
identification of circRNA-disease associations based on learning to rank
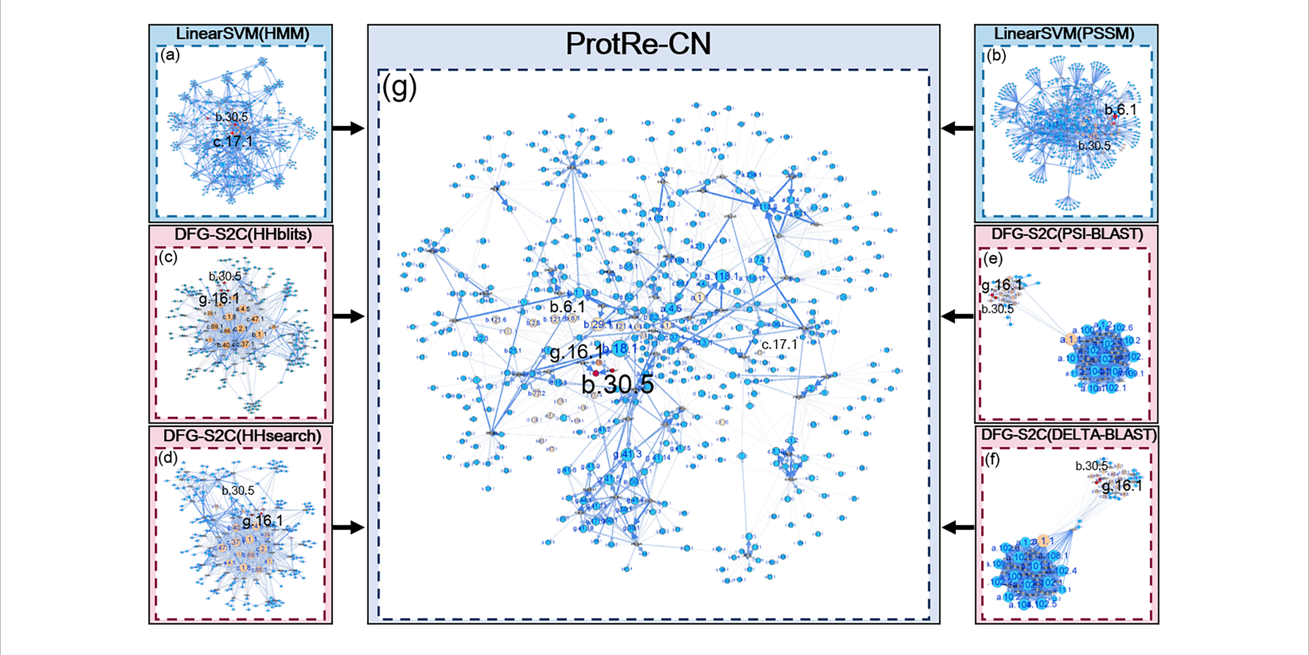
ProtRe-CN
Protein remote homology detection by combining classification methods and network methods via Learning to Rank
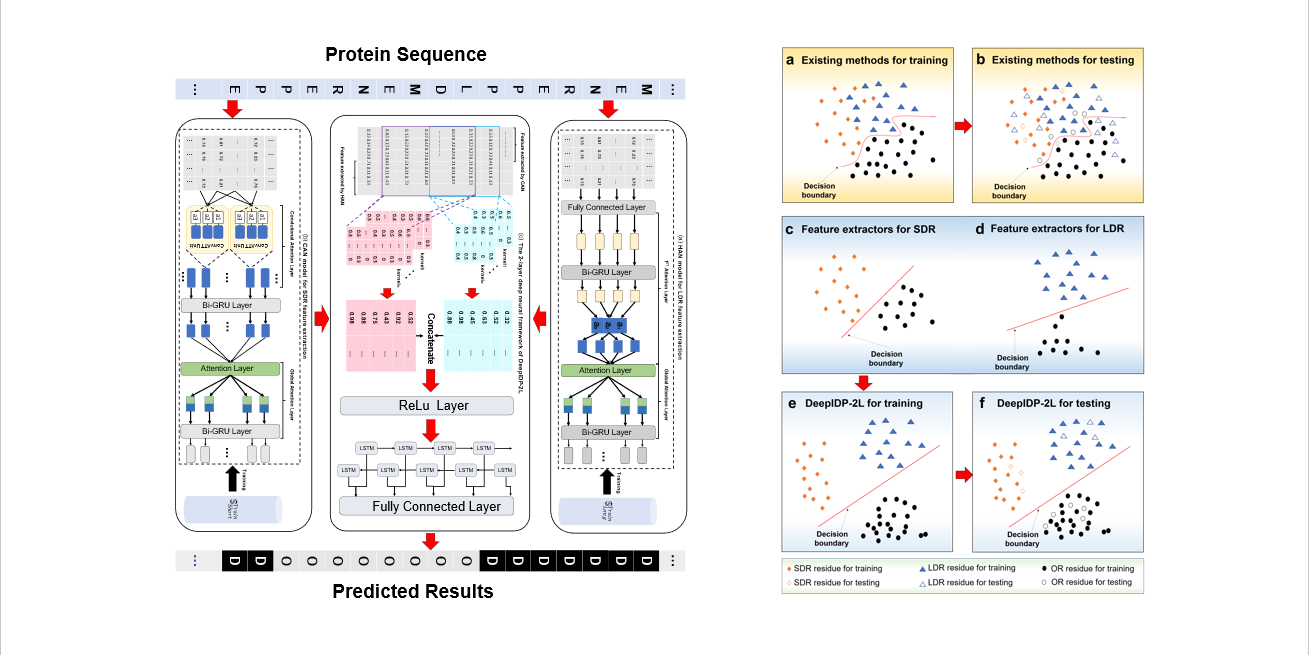
DeepIDP-2L
protein intrinsically disordered region prediction by combining convolutional attention network and hierarchical attention network.
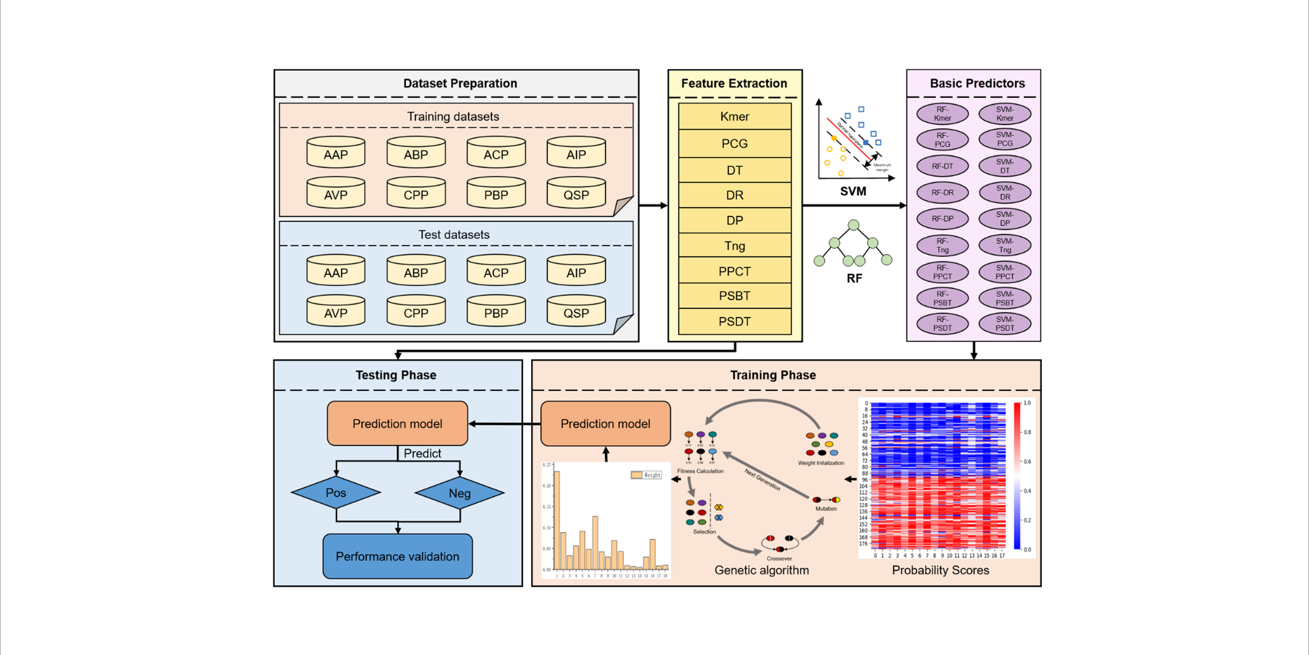
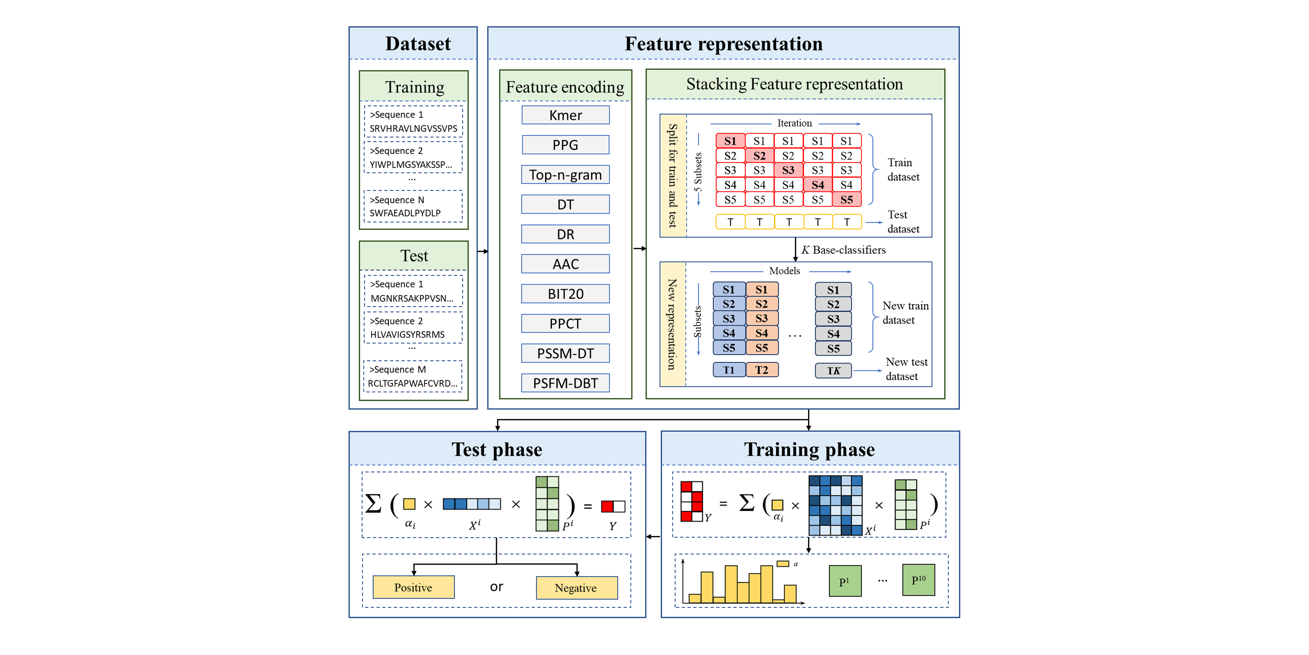
PreTP-Stack
Therapeutic peptides prediction based on auto-weighted multi-view learning
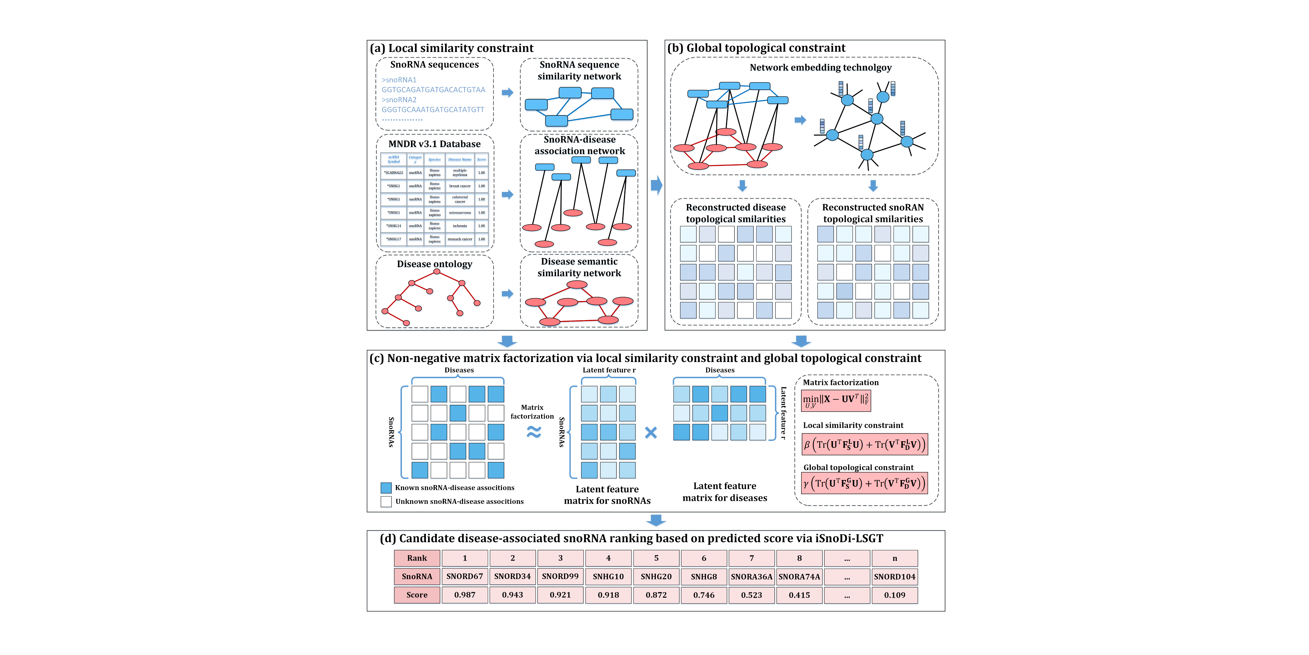
iSnoDi-LSGT
identifying snoRNA-disease associations based on local similarity constraint and global topological constraint
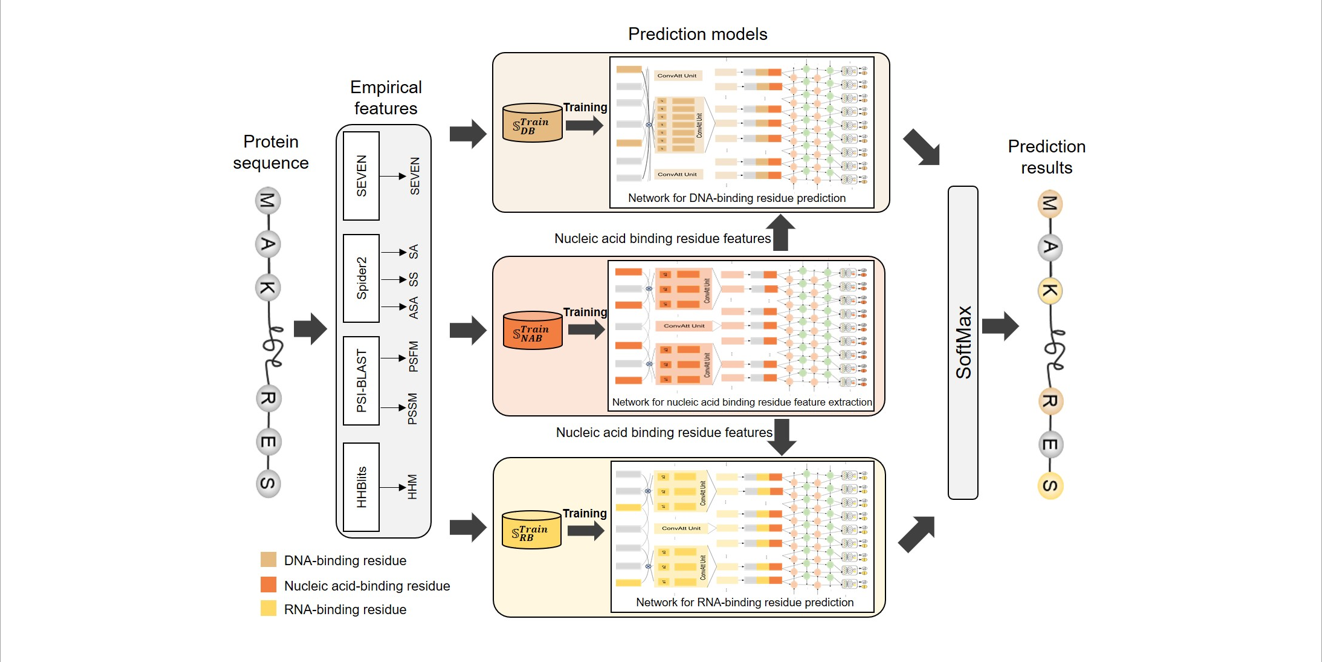
iDRNA-ITF
Identifying DNA- and RNA-binding residues in proteins based on induction and transfer framework
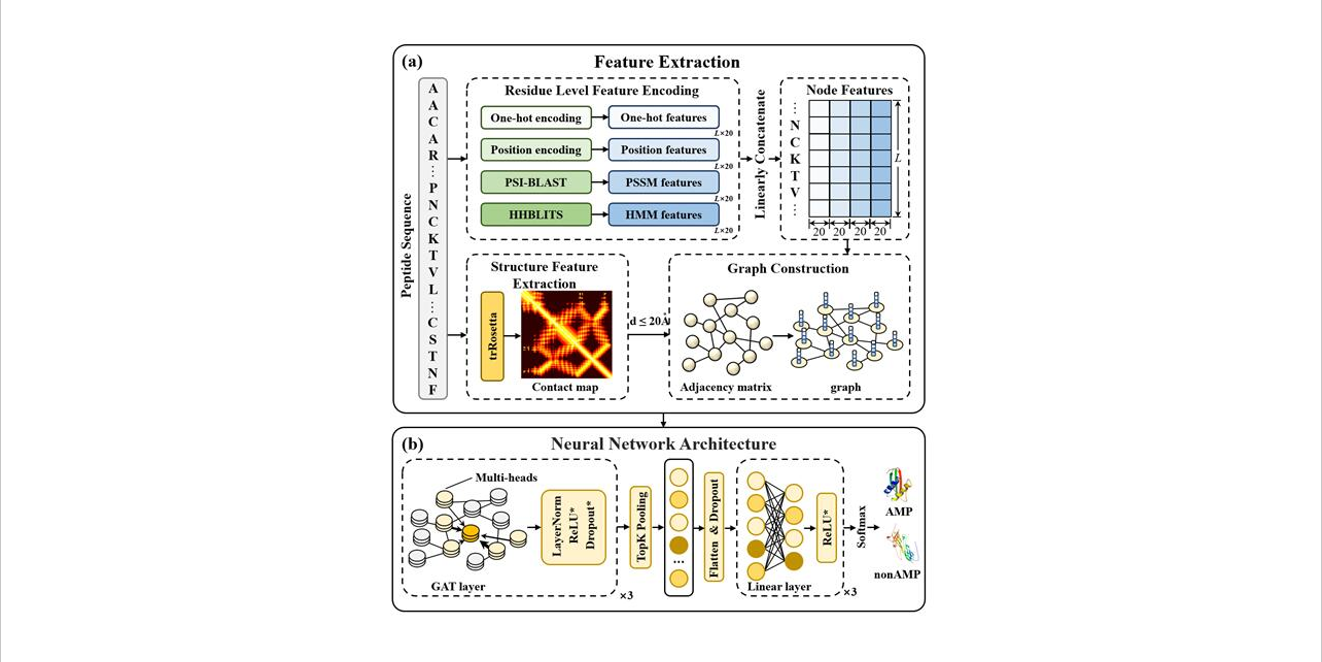
sAMPpred-GAT
Prediction of Antimicrobial Peptides based on Graph Attention Network
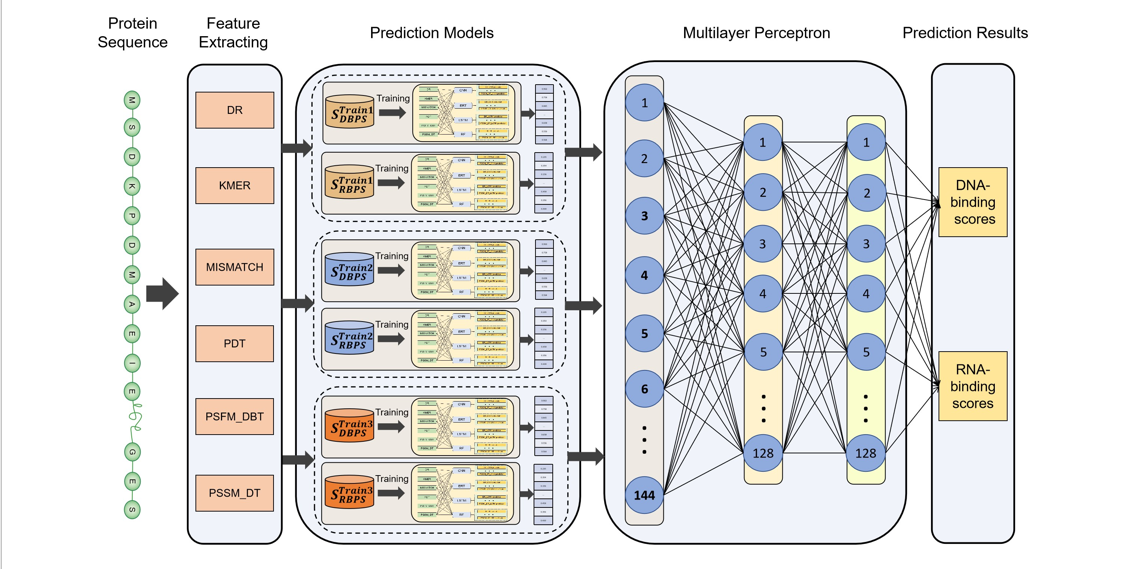
iDRBP-ECHF
Identifying DNA- and RNA- binding proteins based on extensible cubic hybrid framework
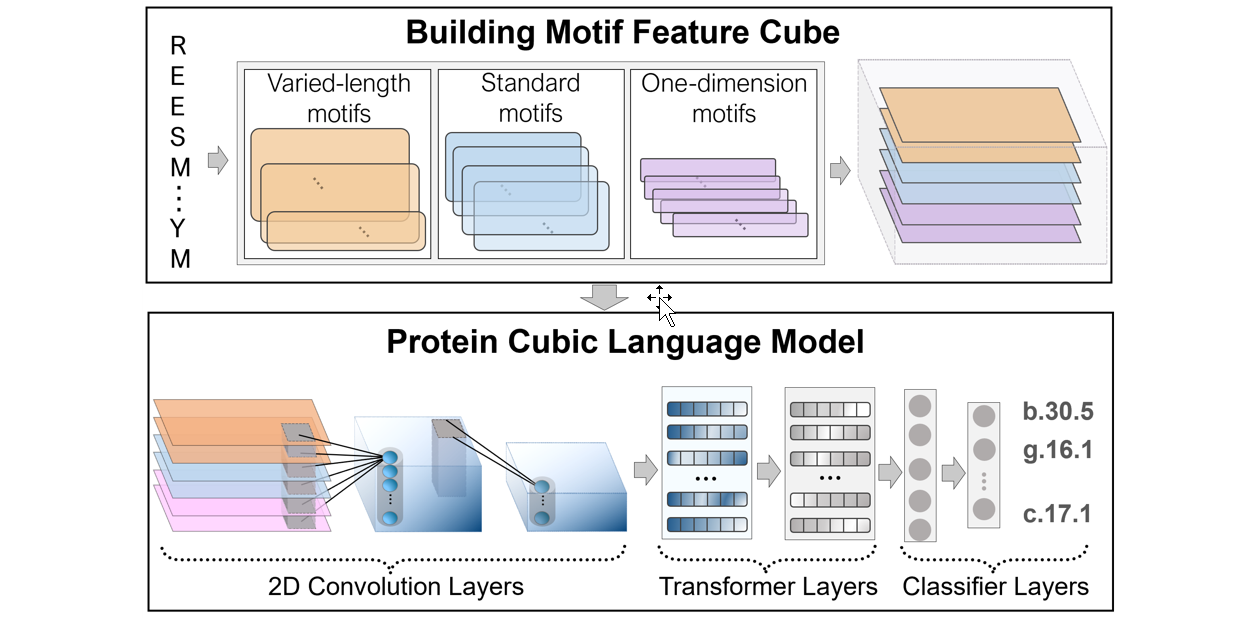
PreHom-PCLM
Protein Remote Homology Detection by Combing Motifs and Protein Cubic Language Model
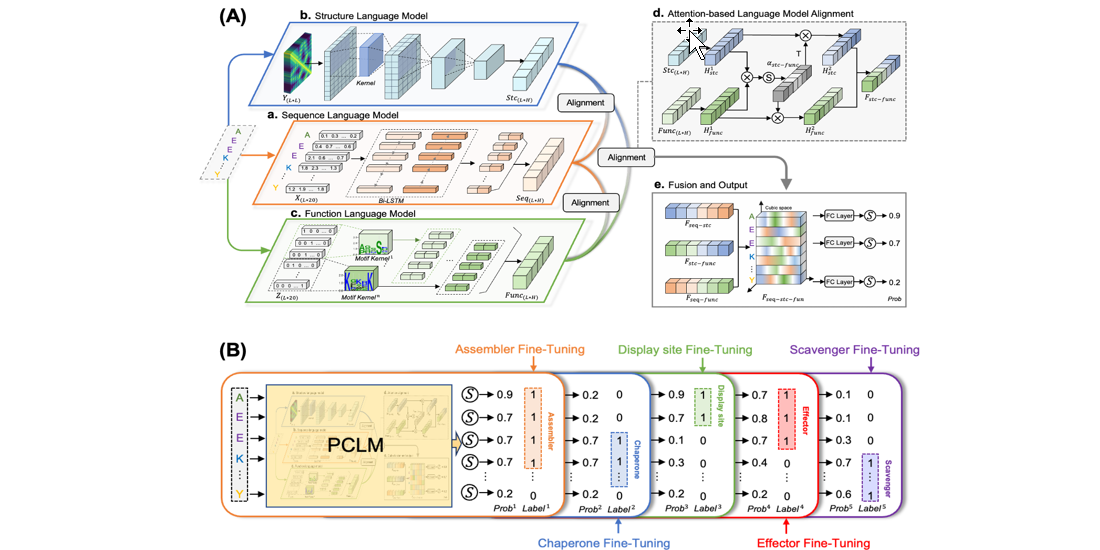
DMFpred
Predicting protein disorder molecular functions based on protein cubic language model
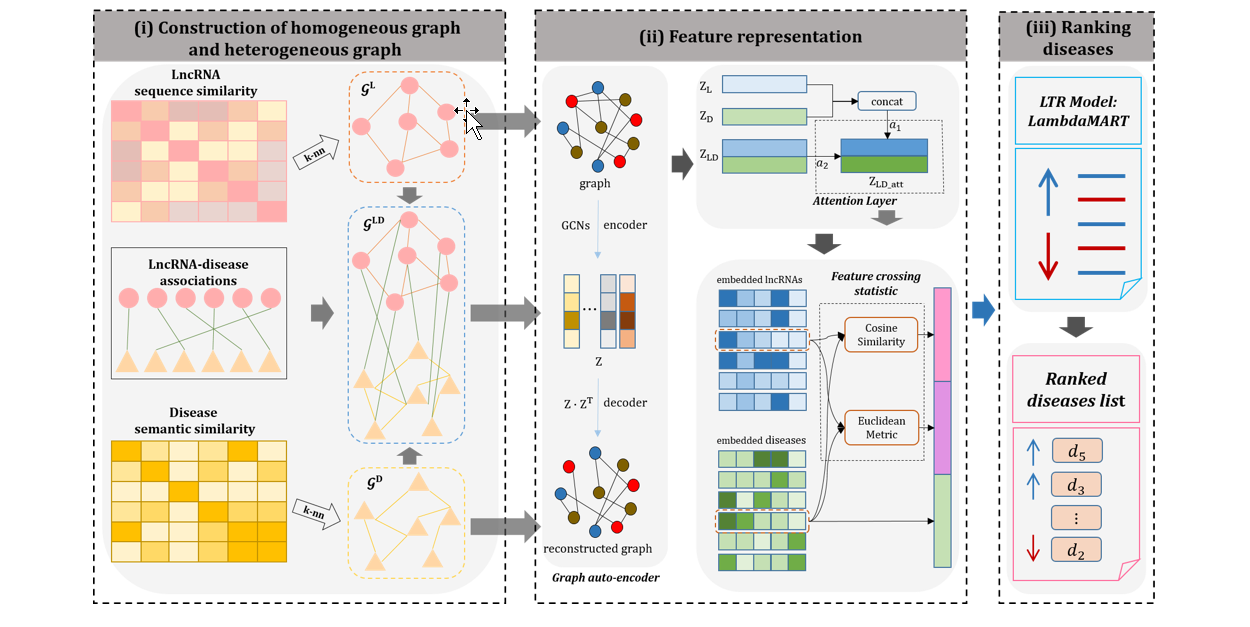
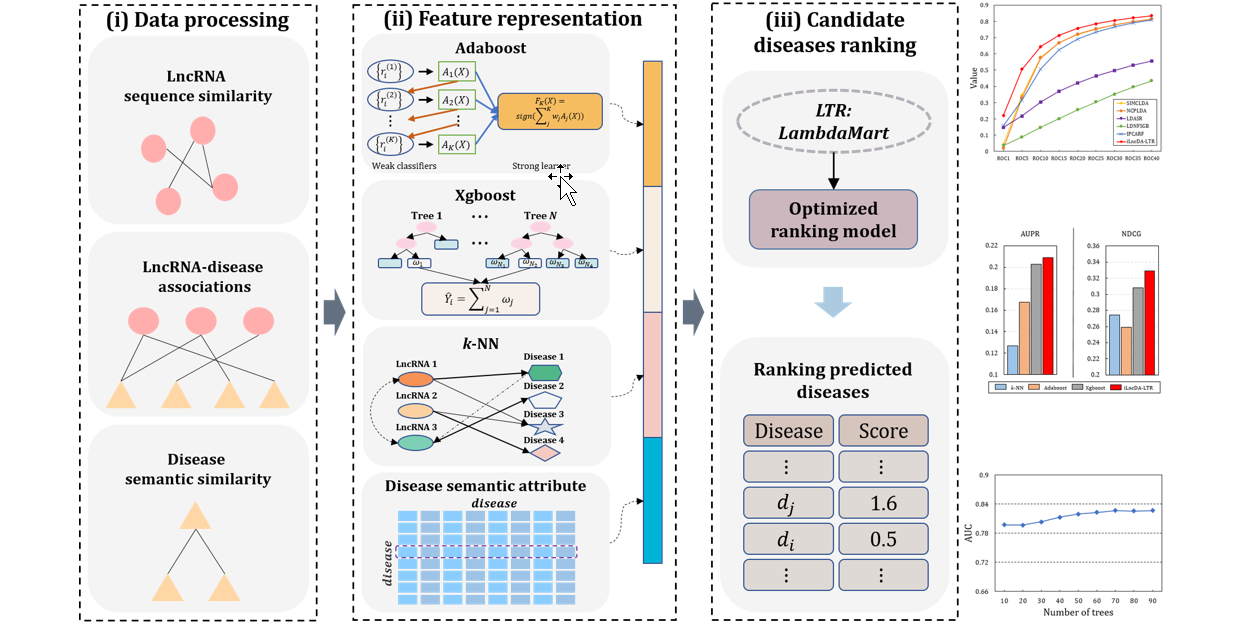
GraLTR-LDA
lncRNA-disease association prediction based on graph auto-encoder and Learning to Rank
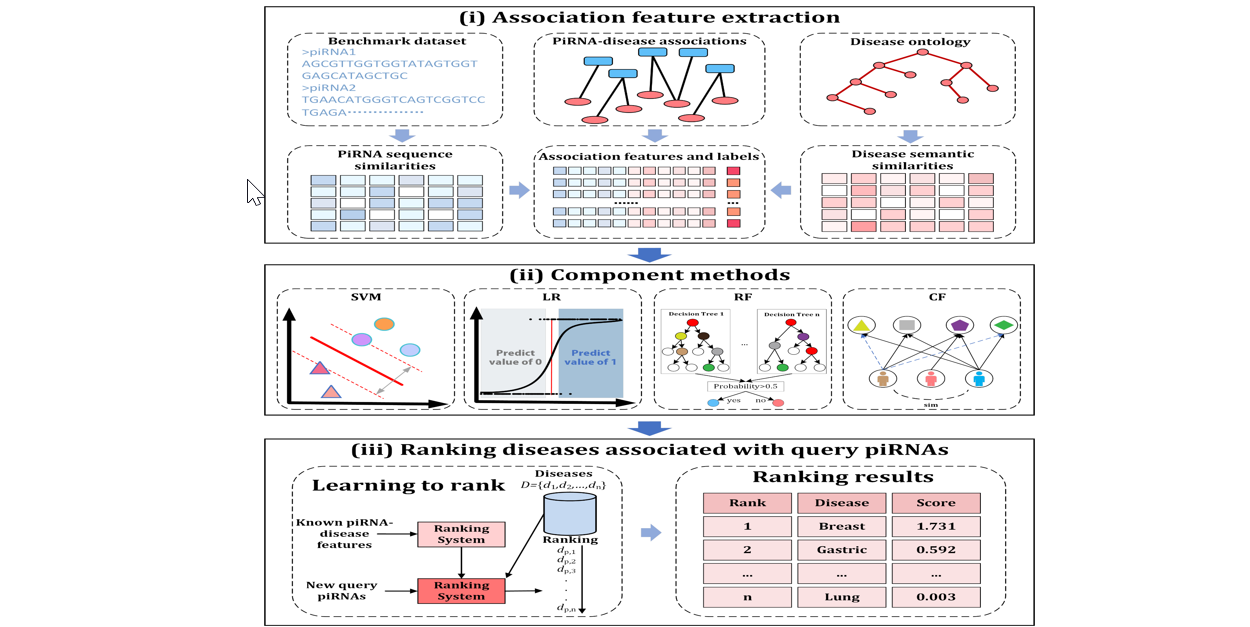
iPiDA-LTR
Identifying piwi-interacting RNA-disease associations based on learning to rank
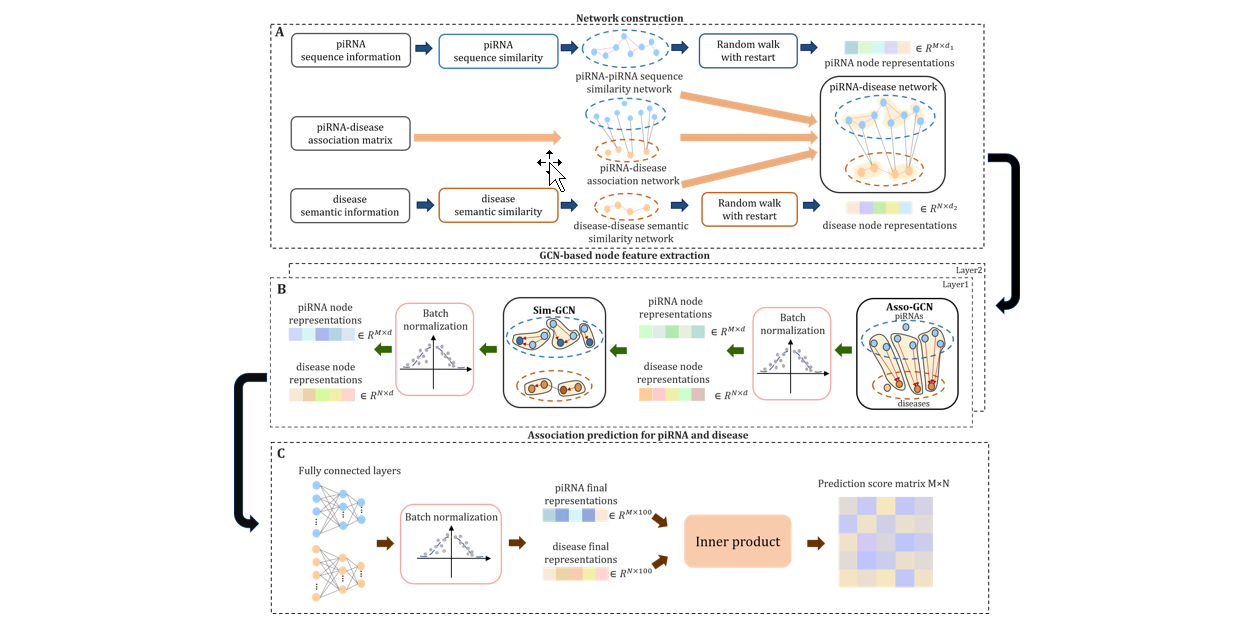
iPiDA-GCN
Identification of piRNA-disease associations based on Graph Convolutional Network

iSnoDi-MDRF
Identifying snoRNA-disease associations based on multiple biological data by ranking framework
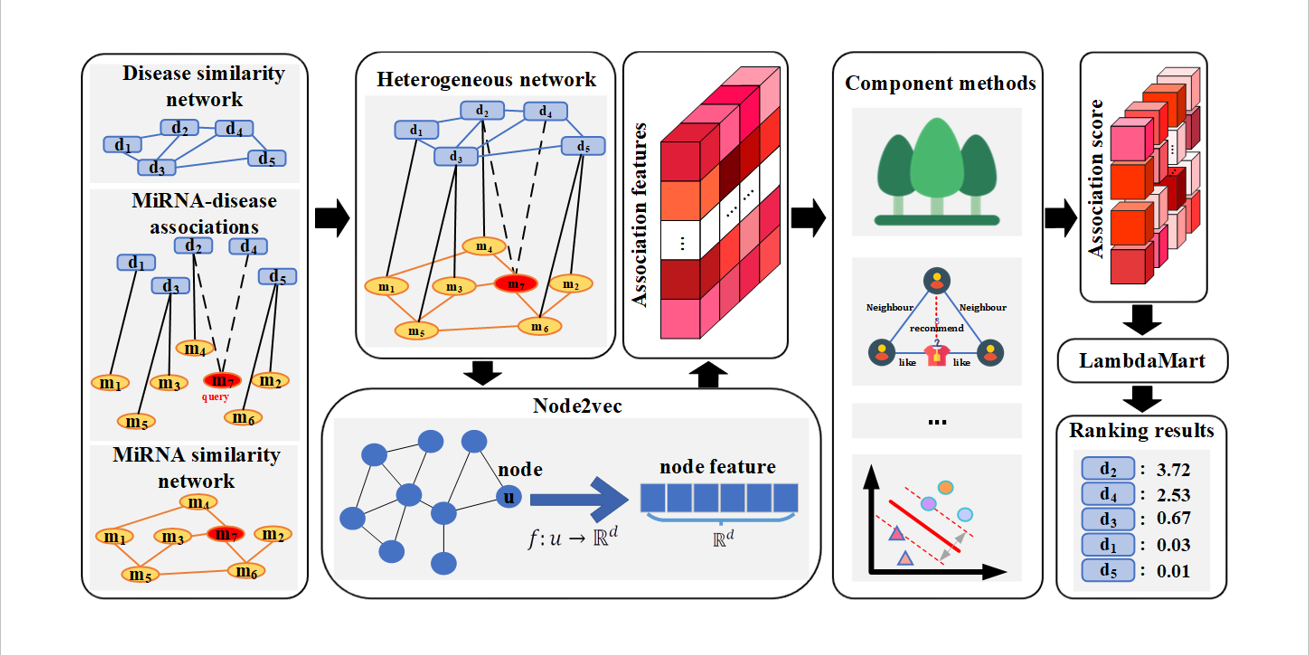
idenMD-NRF
A novel ranking framework for improving the identification of miRNA-disease association
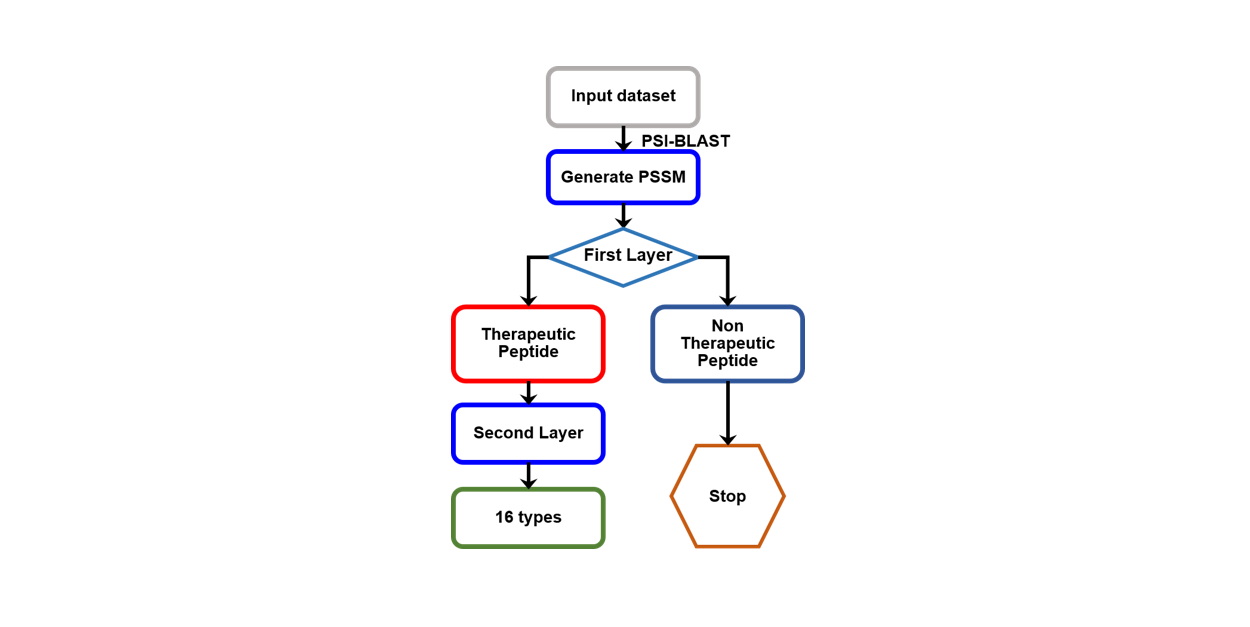
PreTP-2L
Identification of therapeutic peptides and their types using two-layer ensemble learning framework
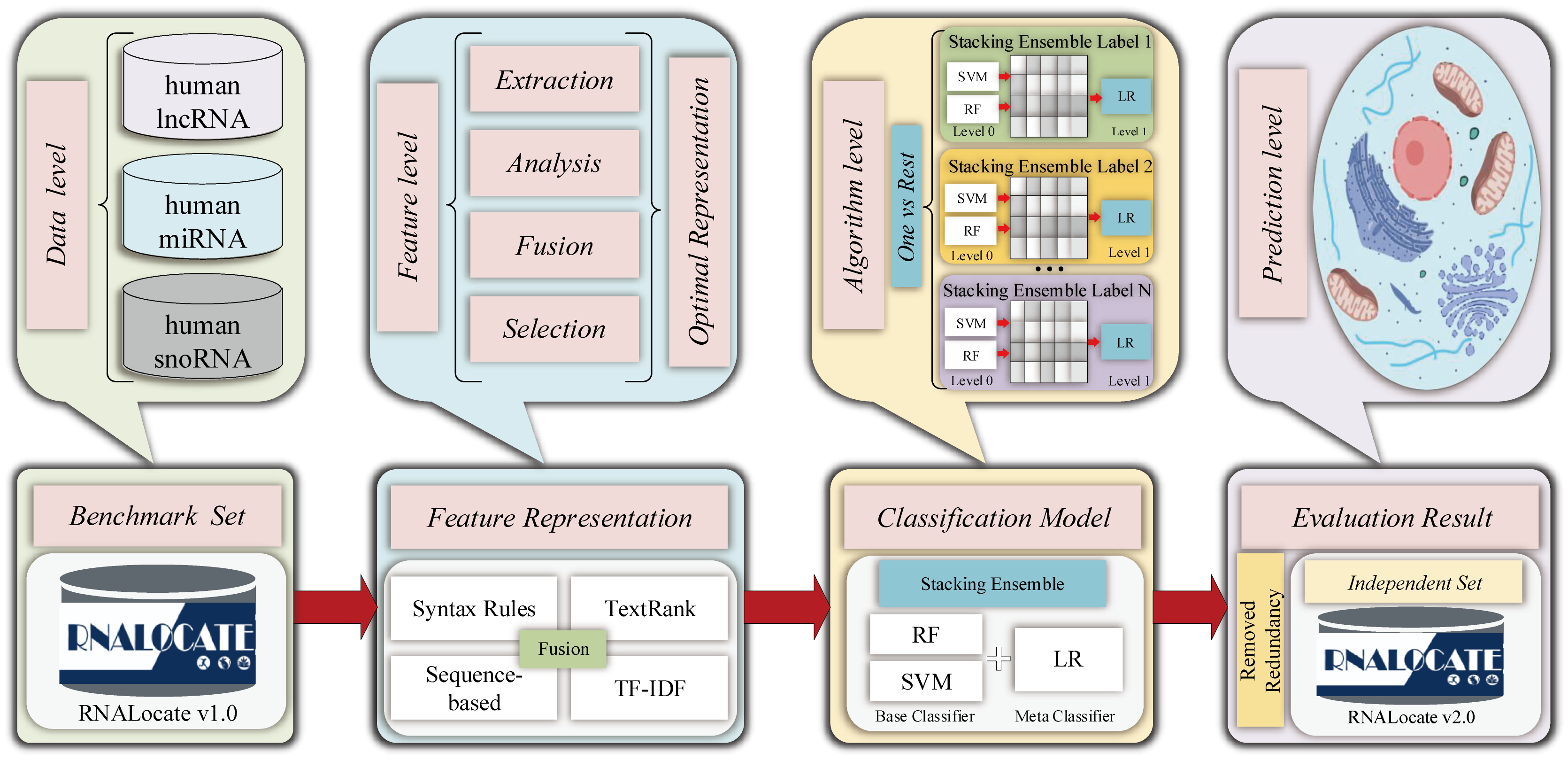
ncRNALocate-EL
A novel multi-label Subcellular Locality prediction model of ncRNA based on ensemble learning

ProFun-SOM
Protein Function Prediction for Specific Ontology based on Multiple Sequence Alignment Reconstruction
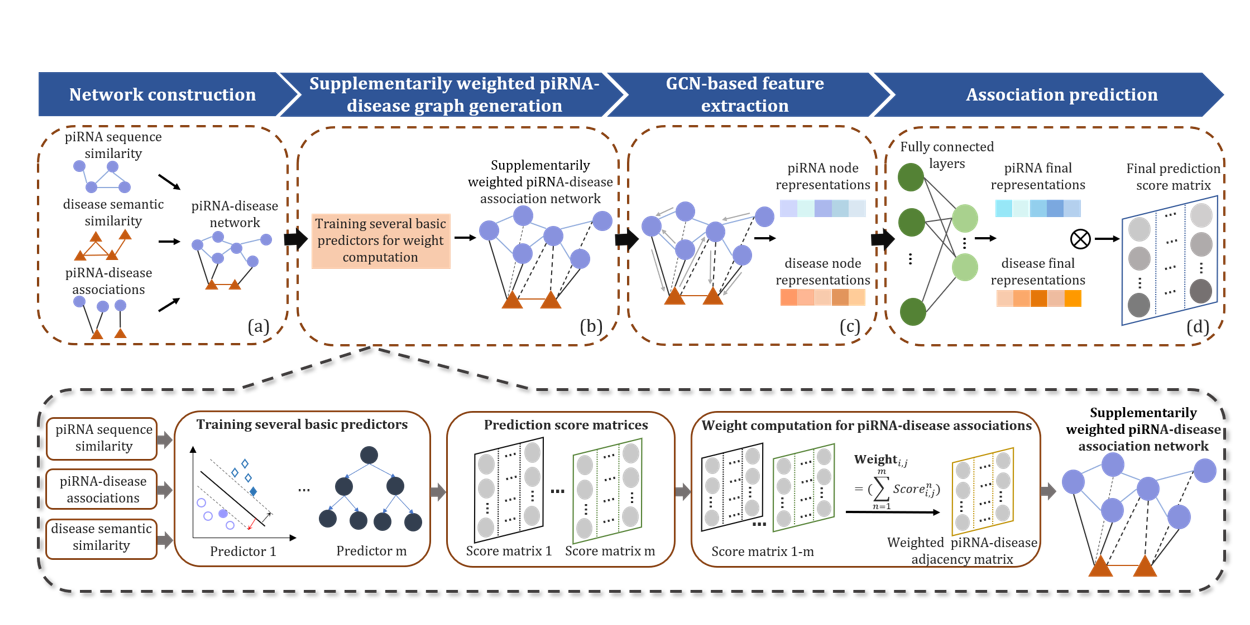
iPiDA-SWGCN
Identification of piRNA-disease associations based on Supplementarily Weighted Graph Convolutional Network
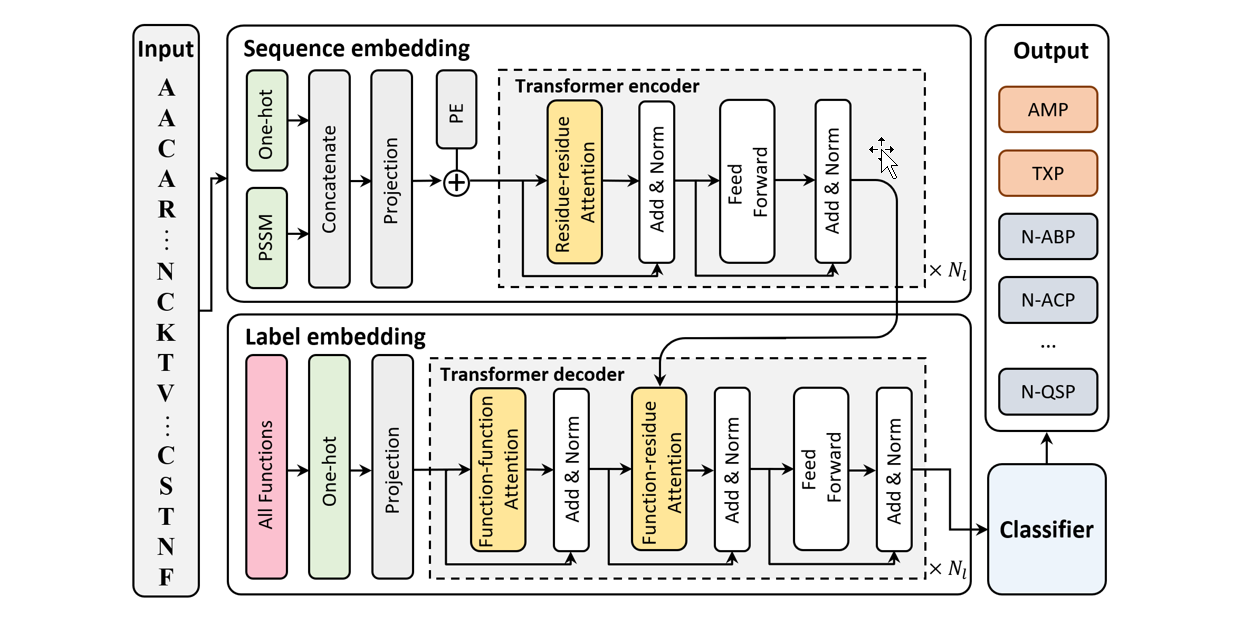
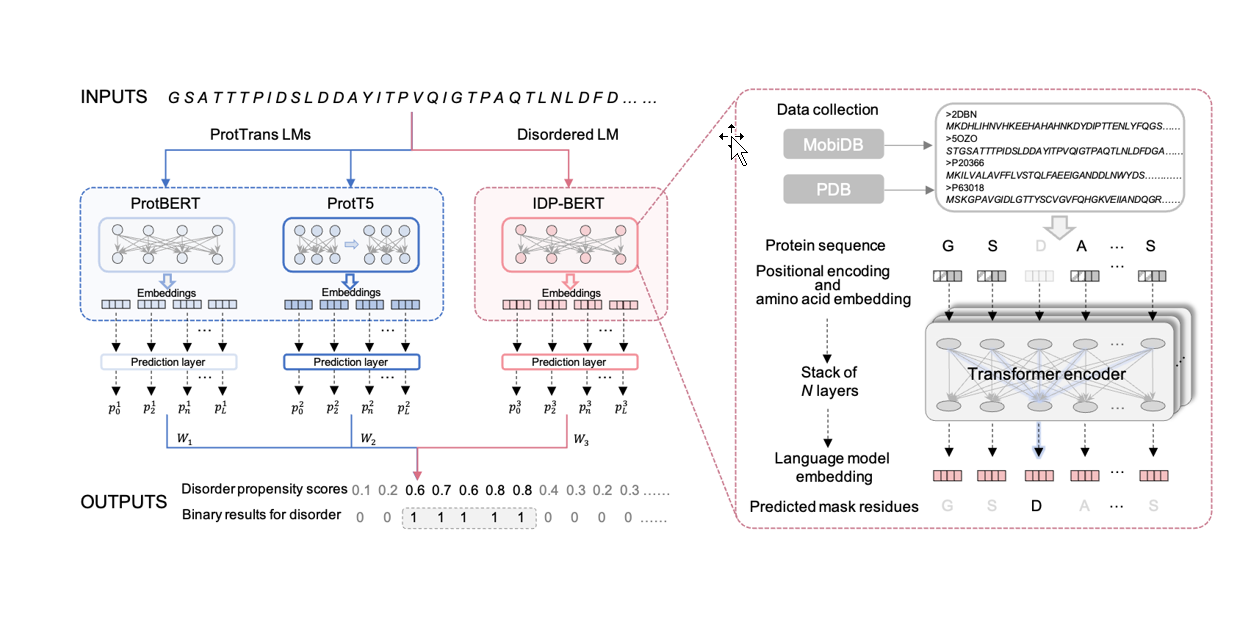
IDP_LM
Prediction of protein intrinsic disorder and disorder functions based on language models
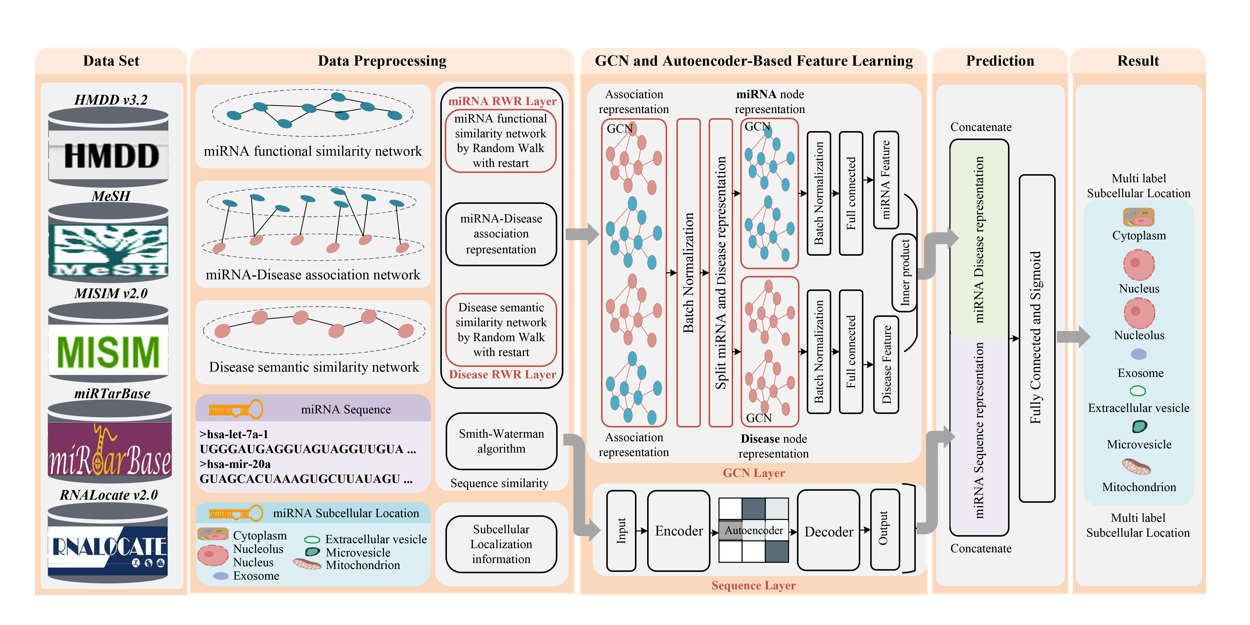
DAmiRLocGNet
miRNA subcellular localization prediction by combining miRNA-disease associations and graph convolutional networks
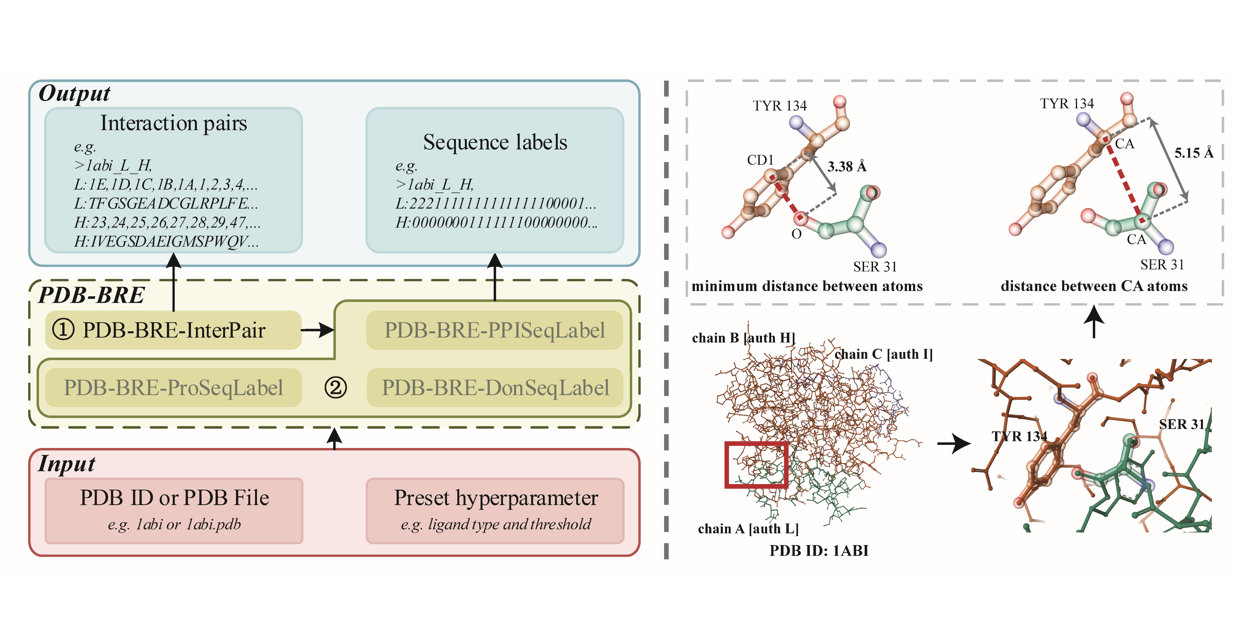
PDB-BRE
A ligand-protein interaction binding residue extractor based on Protein Data Bank
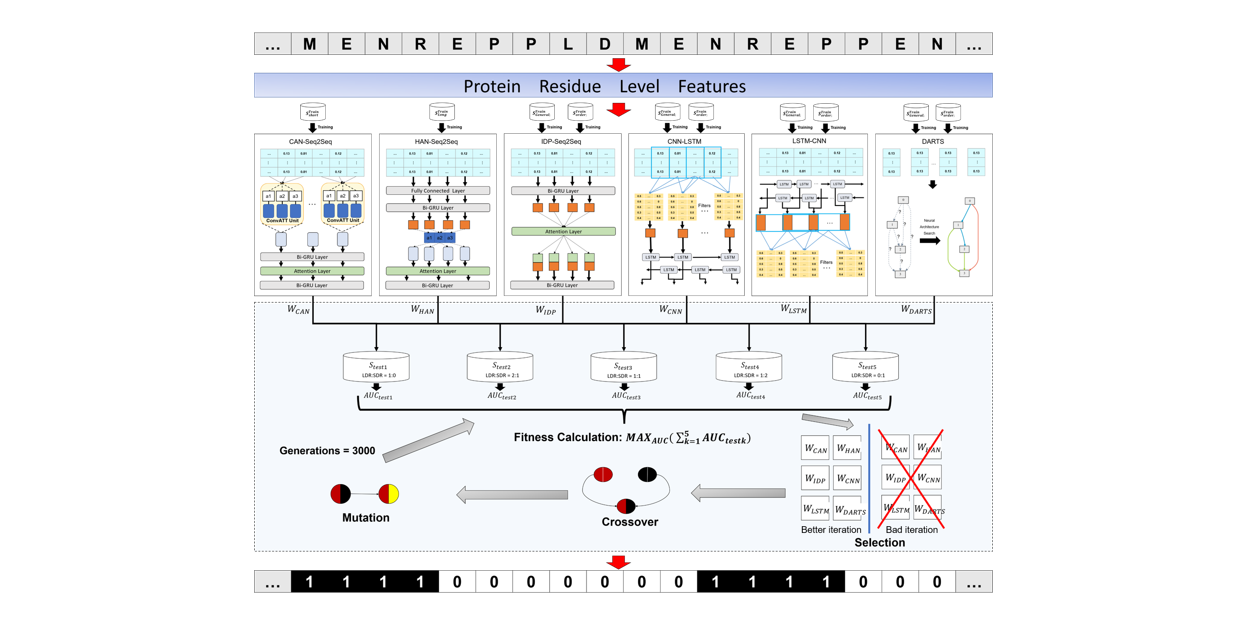
IDP-Fusion
Protein intrinsically disordered region prediction by combining Neural Architecture Search and Multi-objective genetic algorithm
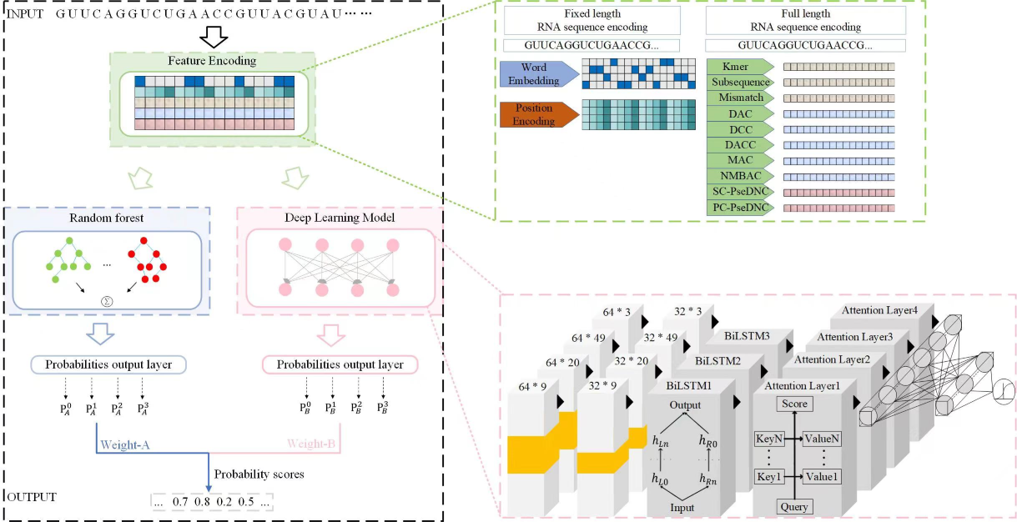
MulStack
An ensemble learning prediction model of multilabel mRNA subcellular localization
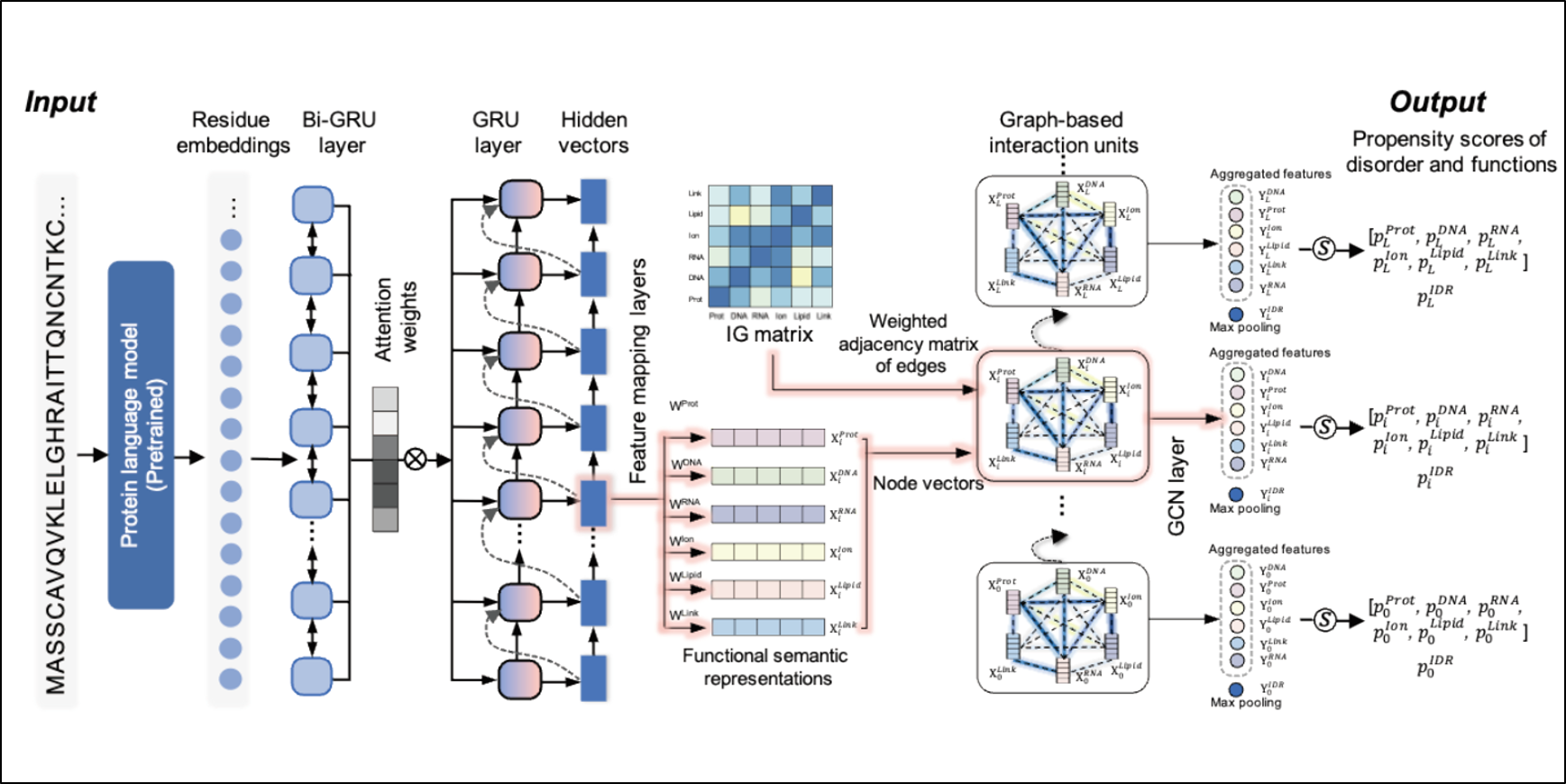
DisoFLAG
Accurate prediction of protein intrinsic disorder and its functions using graph-based interaction protein language model
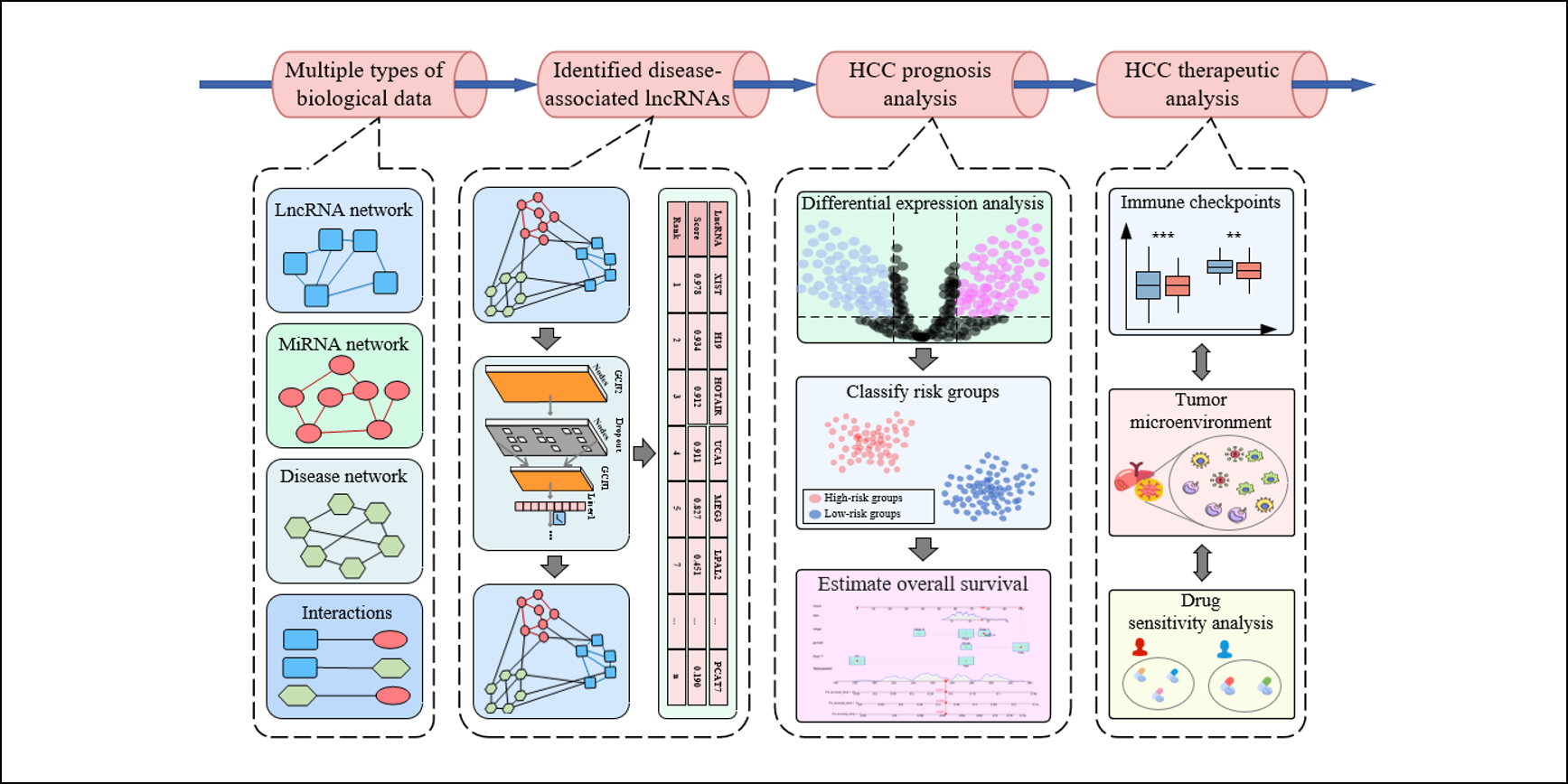
iLncDA-PT
Navigating the LncRNA-disease Pipeline: from Disease-associated LncRNA Identification to Prognosis and Therapeutic for Diseases
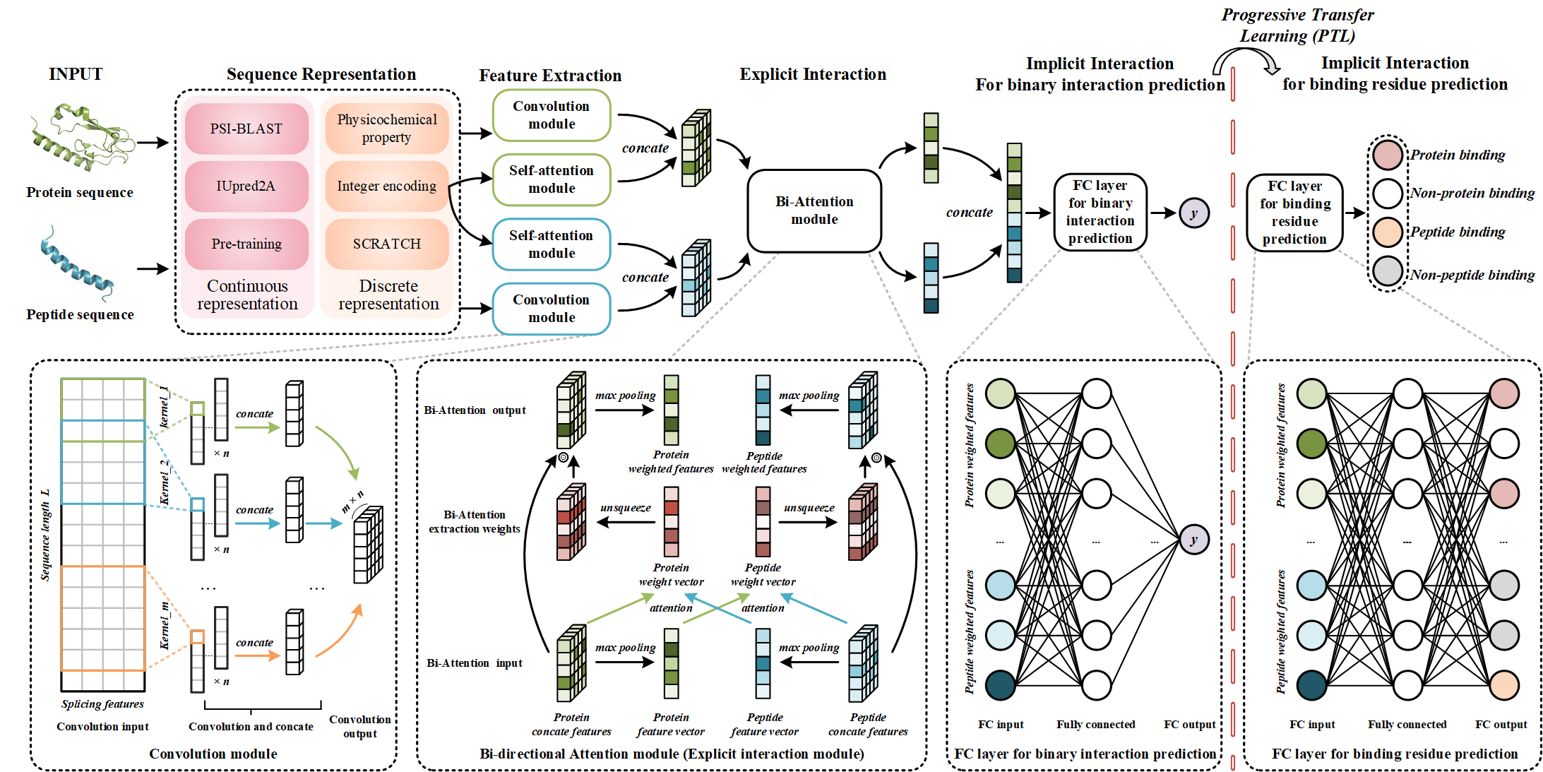
IIDL-PepPI
Peptide-Protein Interaction Profiling Model Based on Interpretable Progressive Transfer Model
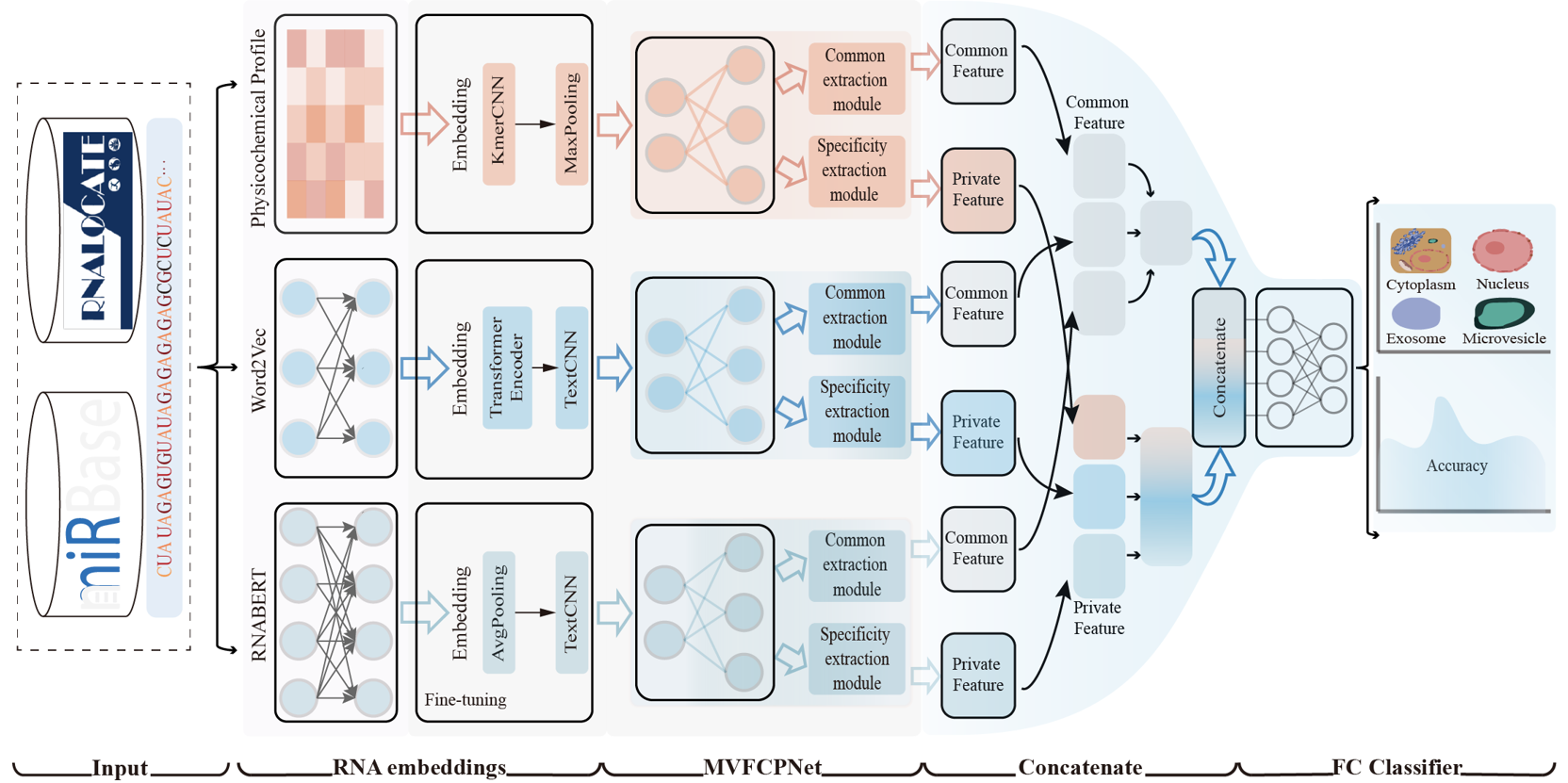
MMLmiRLocNet
A Multi-view Multi-label Learning Approach for miRNA Subcellular Localization Prediction
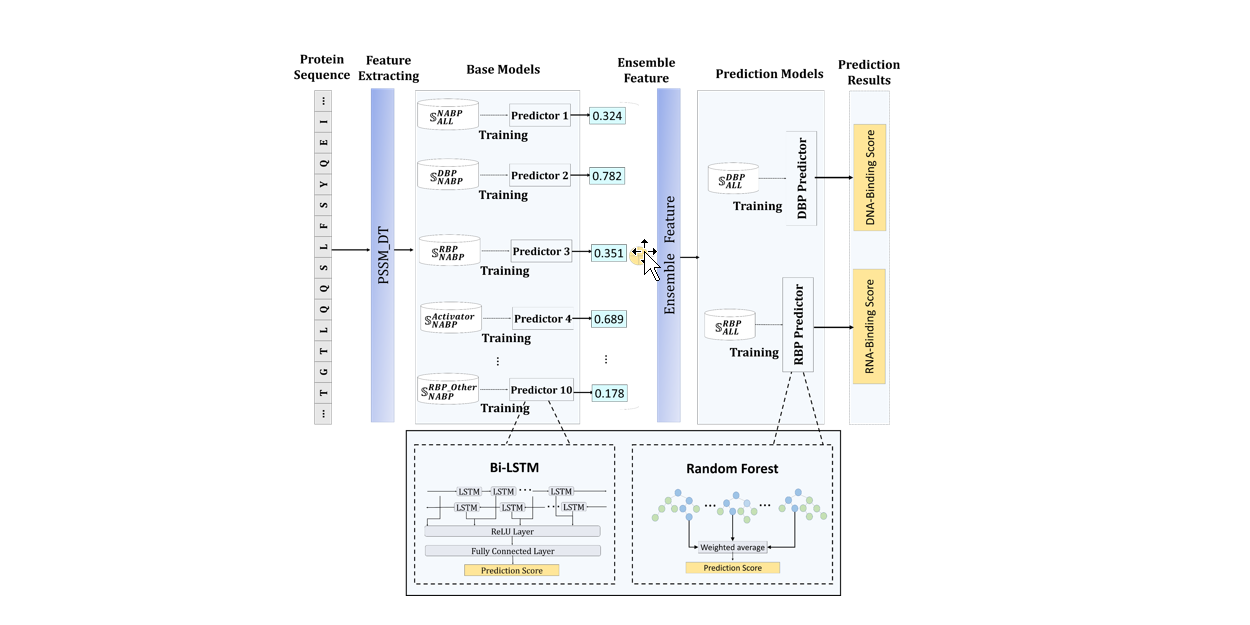
iDRPro-SC
Identifying DNA-Binding Proteins and RNA-Binding Proteins based on Subfunction Classifiers
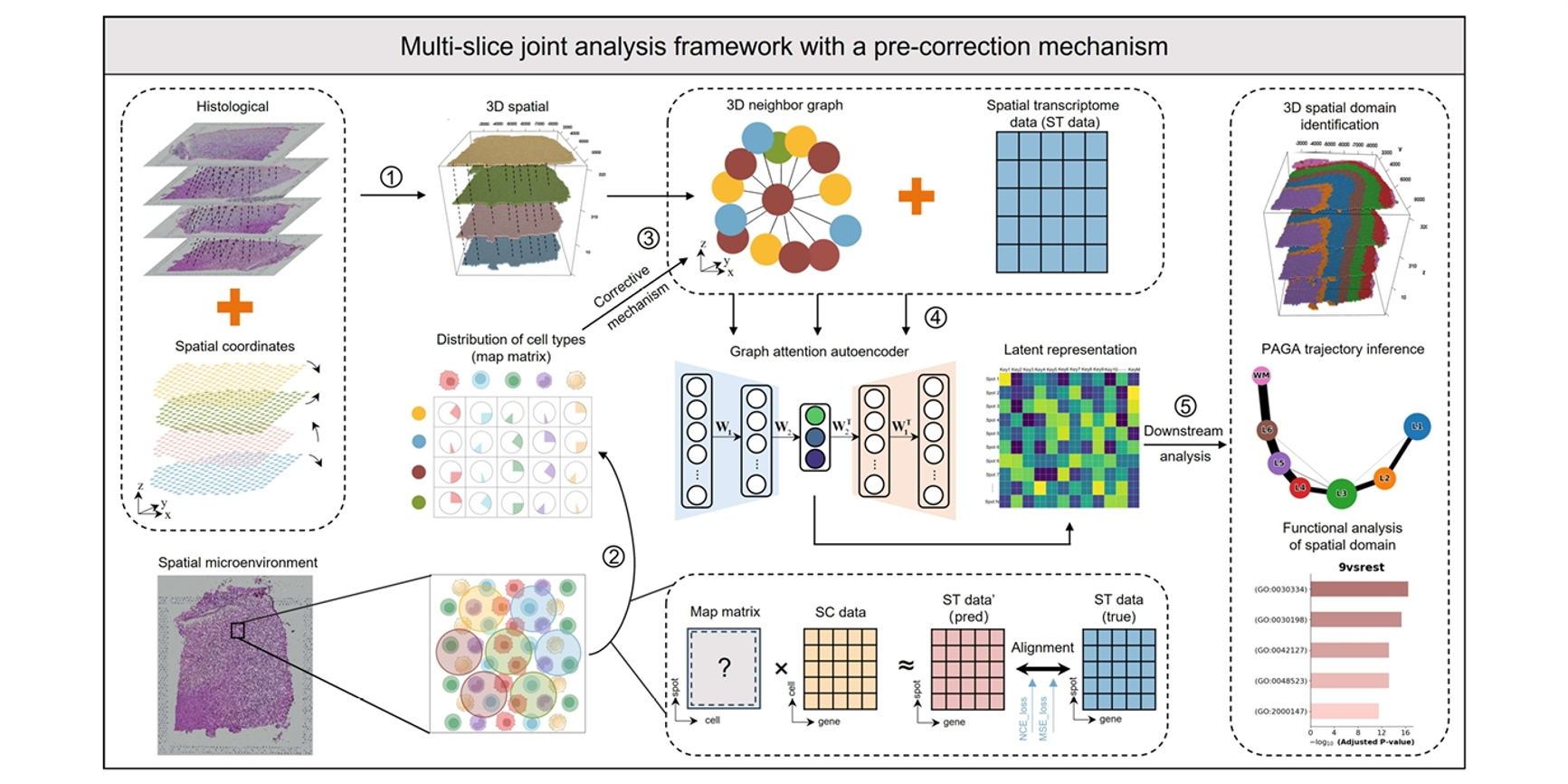
STMSC
A Novel Multi-Slice Framework for Precision 3D Spatial Domain Reconstruction and Disease Pathology Analysis
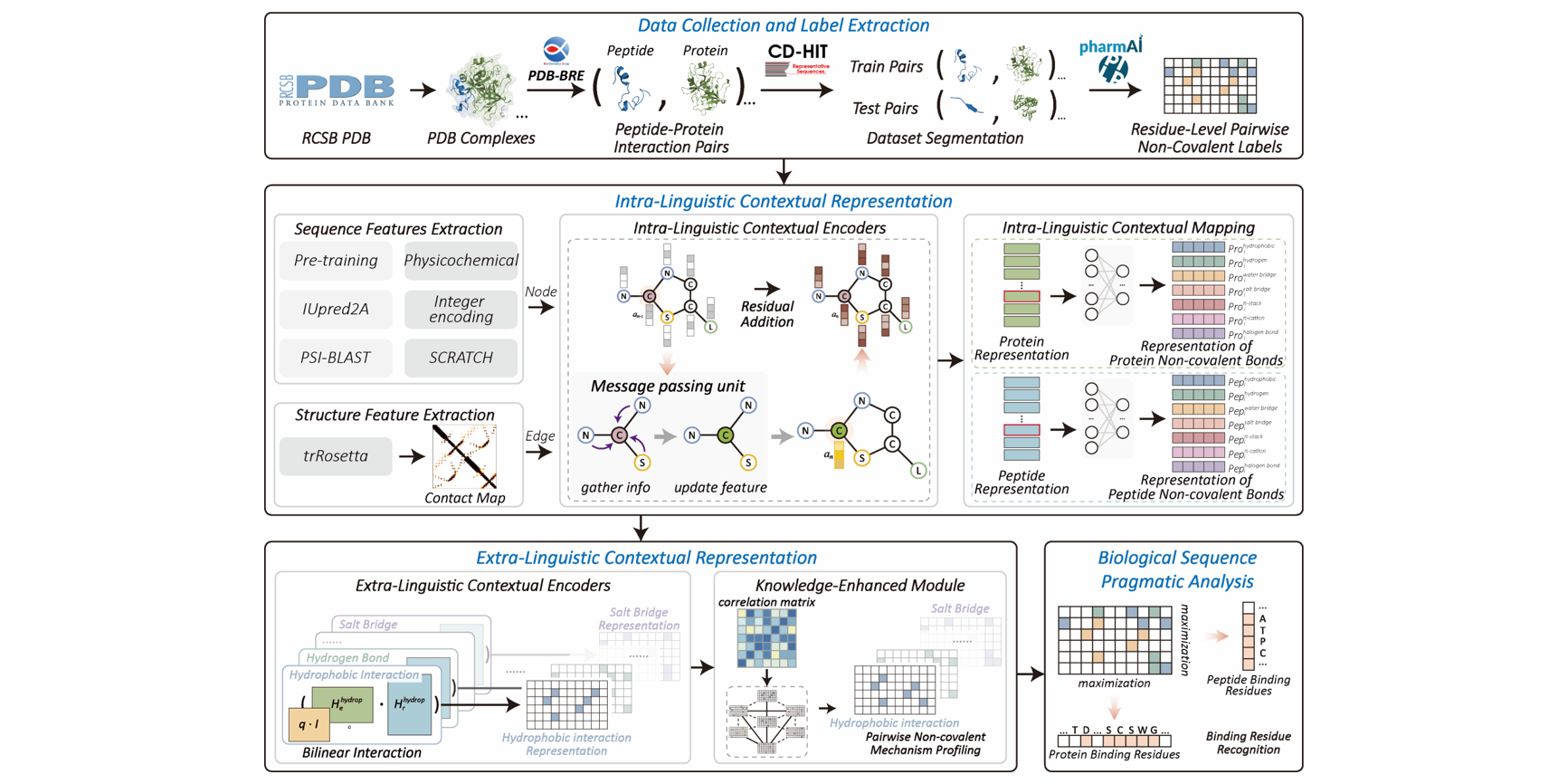
KEIPA
Knowledge-Enhanced Interpretable Pragmatic Analysis for Uncovering Peptide-Protein Pairwise Non-Covalent Mechanisms
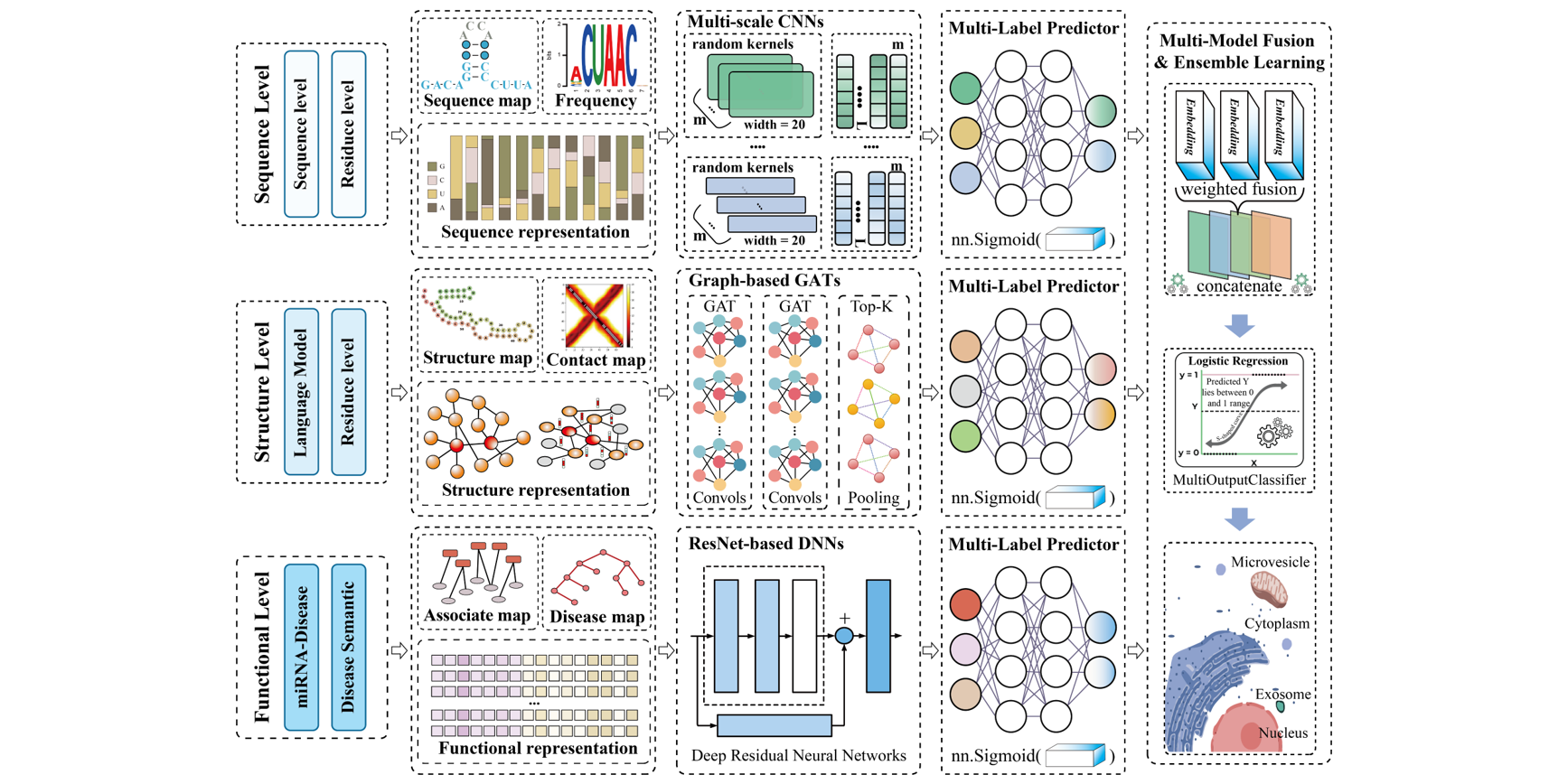
MMFmiRLocEL
A Multi-model Fusion and Ensemble Learning Approach for Identifying miRNA Subcellular Localization using RNA Structure Language Model
Contact
Please constact us by email
Email Us
Prof. Dr. Bin Liu, email: bliu@bliulab.net
